Before we Start
Overview
Teaching: 10 min
Exercises: 5 minQuestions
What have I forgotten about R and RStudio?
How to interact with R?
How to manage your environment?
How to install packages?
Objectives
Install latest version of R.
Install latest version of RStudio.
Navigate the RStudio GUI.
Install additional packages using the packages tab.
Install additional packages using R code.
What is R? What is RStudio?
The term “R” is used to refer to both the programming language and the
software that interprets the scripts written using it.
RStudio is currently a very popular way to not only write your R scripts but also to interact with the R software. To function correctly, RStudio needs R and therefore both need to be installed on your computer.
To make it easier to interact with R, we will use RStudio. RStudio is the most popular IDE (Integrated Development Environmemt) for R. An IDE is a piece of software that provides tools to make programming easier.
Why learn R?
R does not involve lots of pointing and clicking, and that’s a good thing
The learning curve might be steeper than with other software, but with R, the results of your analysis do not rely on remembering a succession of pointing and clicking, but instead on a series of written commands, and that’s a good thing! So, if you want to redo your analysis because you collected more data, you don’t have to remember which button you clicked in which order to obtain your results; you just have to run your script again.
Working with scripts makes the steps you used in your analysis clear, and the code you write can be inspected by someone else who can give you feedback and spot mistakes.
Working with scripts forces you to have a deeper understanding of what you are doing, and facilitates your learning and comprehension of the methods you use.
R code is great for reproducibility
Reproducibility is when someone else (including your future self) can obtain the same results from the same dataset when using the same analysis.
R integrates with other tools to generate manuscripts from your code. If you collect more data, or fix a mistake in your dataset, the figures and the statistical tests in your manuscript are updated automatically.
An increasing number of journals and funding agencies expect analyses to be reproducible, so knowing R will give you an edge with these requirements.
R is interdisciplinary and extensible
With 18,000+ packages that can be installed to extend its capabilities, R provides a framework that allows you to combine statistical approaches from many scientific disciplines to best suit the analytical framework you need to analyze your data. For instance, R has packages for image analysis, GIS, time series, population genetics, and a lot more.
R works on data of all shapes and sizes
The skills you learn with R scale easily with the size of your dataset. Whether your dataset has hundreds or millions of lines, it won’t make much difference to you.
R is designed for data analysis. It comes with special data structures and data types that make handling of missing data and statistical factors convenient.
R can connect to spreadsheets, databases, and many other data formats, on your computer or on the web.
R produces high-quality graphics
The plotting functionalities in R are endless, and allow you to adjust any aspect of your graph to convey most effectively the message from your data.
R has a large and welcoming community
Thousands of people use R daily. Many of them are willing to help you through mailing lists and websites such as Stack Overflow, or on the RStudio community. Questions which are backed up with short, reproducible code snippets are more likely to attract knowledgeable responses.
Not only is R free, but it is also open-source and cross-platform
Anyone can inspect the source code to see how R works. Because of this transparency, there is less chance for mistakes, and if you (or someone else) find some, you can report and fix bugs.
Because R is open source and is supported by a large community of developers and users, there is a very large selection of third-party add-on packages which are freely available to extend R’s native capabilities.


A tour of RStudio
Knowing your way around RStudio
Let’s start by learning about RStudio, which is an Integrated Development Environment (IDE) for working with R.
The RStudio IDE open-source product is free under the Affero General Public License (AGPL) v3. The RStudio IDE is also available with a commercial license and priority email support from RStudio, Inc.
We will use the RStudio IDE to write code, navigate the files on our computer, inspect the variables we create, and visualize the plots we generate. RStudio can also be used for other things (e.g., version control, developing packages, writing Shiny apps) that we will not cover during the workshop.
One of the advantages of using RStudio is that all the information you need to write code is available in a single window. Additionally, RStudio provides many shortcuts, autocompletion, and highlighting for the major file types you use while developing in R. RStudio makes typing easier and less error-prone.
Getting set up
It is good practice to keep a set of related data, analyses, and text self-contained in a single folder called the working directory. All of the scripts within this folder can then use relative paths to files. Relative paths indicate where inside the project a file is located (as opposed to absolute paths, which point to where a file is on a specific computer). Working this way makes it a lot easier to move your project around on your computer and share it with others without having to directly modify file paths in the individual scripts.
RStudio provides a helpful set of tools to do this through its “Projects” interface, which not only creates a working directory for you but also remembers its location (allowing you to quickly navigate to it). The interface also (optionally) preserves custom settings and open files to make it easier to resume work after a break.
Create a new project
- Under the
Filemenu, click onNew project, chooseNew directory, thenNew project - Enter a name for this new folder (or “directory”) and choose a convenient
location for it. This will be your working directory for the rest of the
day (e.g.,
~/data-carpentry) - Click on
Create project - Create a new file where we will type our scripts. Go to File > New File > R
script. Click the save icon on your toolbar and save your script as
“
script.R”.
The simplest way to open an RStudio project once it has been created is to
navigate through your files to where the project was saved and double
click on the .Rproj (blue cube) file. This will open RStudio and start your R
session in the same directory as the .Rproj file. All your data, plots and
scripts will now be relative to the project directory. RStudio projects have the
added benefit of allowing you to open multiple projects at the same time each
open to its own project directory. This allows you to keep multiple projects
open without them interfering with each other.
The RStudio Interface
Let’s take a quick tour of RStudio.
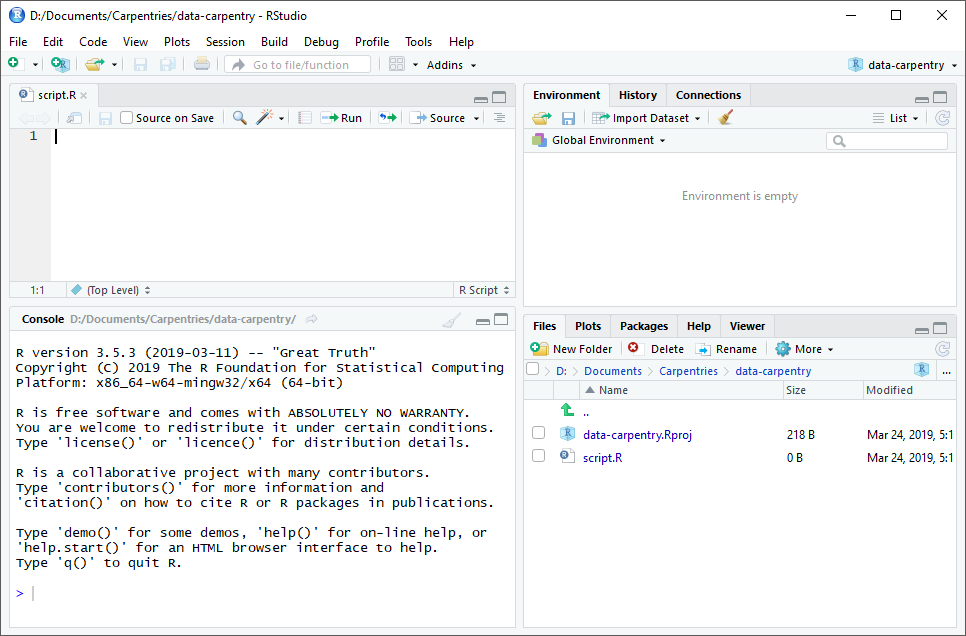
RStudio is divided into four “panes”. The placement of these panes and their content can be customized (see menu, Tools -> Global Options -> Pane Layout).
The Default Layout is:
- Top Left - Source: your scripts and documents
- Bottom Left - Console: what R would look and be like without RStudio
- Top Right - Enviornment/History: look here to see what you have done
- Bottom Right - Files and more: see the contents of the project/working directory here, like your Script.R file
Organizing your working directory
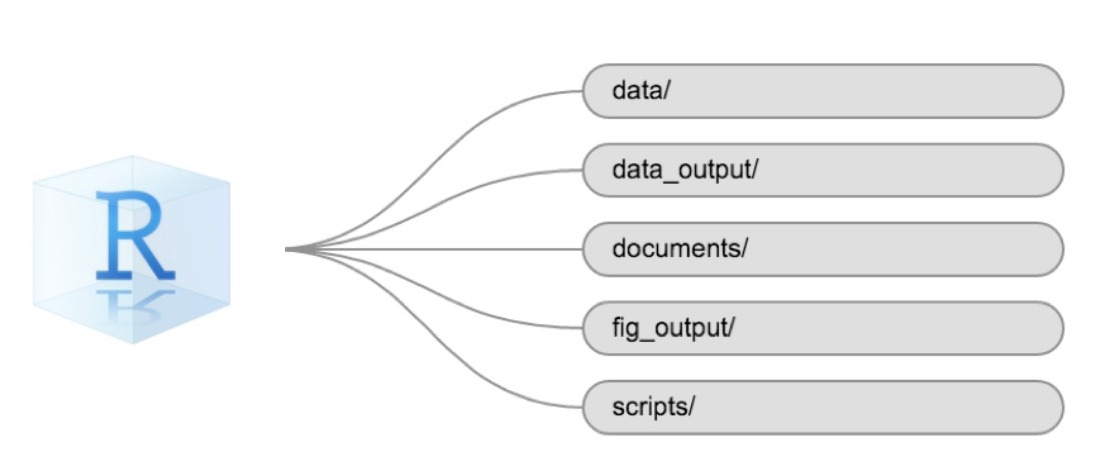
Using a consistent folder structure across your projects will help keep things organized and make it easy to find/file things in the future. This can be especially helpful when you have multiple projects. In general, you might create directories (folders) for scripts, data, and documents. Here are some examples of suggested directories:
data/Use this folder to store your raw data and intermediate datasets. For the sake of transparency and provenance, you should always keep a copy of your raw data accessible and do as much of your data cleanup and preprocessing programmatically (i.e., with scripts, rather than manually) as possible.data_output/When you need to modify your raw data, it might be useful to store the modified versions of the datasets in a different folder.documents/Used for outlines, drafts, and other text.fig_output/This folder can store the graphics that are generated by your scripts.scripts/A place to keep your R scripts for different analyses or plotting.
You may want additional directories or subdirectories depending on your project needs, but these should form the backbone of your working directory.
The working directory
The working directory is an important concept to understand. It is the place where R will look for and save files. When you write code for your project, your scripts should refer to files in relation to the root of your working directory and only to files within this structure.
Using RStudio projects makes this easy and ensures that your working directory
is set up properly. If you need to check it, you can use getwd(). If for some
reason your working directory is not the same as the location of your RStudio
project, it is likely that you opened an R script or RMarkdown file not your
.Rproj file. You should close out of RStudio and open the .Rproj file by
double clicking on the blue cube!
Interacting with R
The basis of programming is that we write down instructions for the computer to follow, and then we tell the computer to follow those instructions. We write, or code, instructions in R because it is a common language that both the computer and we can understand. We call the instructions commands and we tell the computer to follow the instructions by executing (also called running) those commands.
There are two main ways of interacting with R: by using the console or by using script files (plain text files that contain your code). The console pane (in RStudio, the bottom left panel) is the place where commands written in the R language can be typed and executed immediately by the computer. It is also where the results will be shown for commands that have been executed. You can type commands directly into the console and press Enter to execute those commands, but they will be forgotten when you close the session.
Because we want our code and workflow to be reproducible, it is better to type the commands we want in the script editor and save the script. This way, there is a complete record of what we did, and anyone (including our future selves!) can easily replicate the results on their computer.
RStudio allows you to execute commands directly from the script editor by using the Ctrl + Enter shortcut (on Mac, Cmd + Return will work). The command on the current line in the script (indicated by the cursor) or all of the commands in selected text will be sent to the console and executed when you press Ctrl + Enter. If there is information in the console you do not need anymore, you can clear it with Ctrl + L. You can find other keyboard shortcuts in this RStudio cheatsheet about the RStudio IDE.
At some point in your analysis, you may want to check the content of a variable or the structure of an object without necessarily keeping a record of it in your script. You can type these commands and execute them directly in the console. RStudio provides the Ctrl + 1 and Ctrl + 2 shortcuts allow you to jump between the script and the console panes.
If R is ready to accept commands, the R console shows a > prompt. If R
receives a command (by typing, copy-pasting, or sent from the script editor using
Ctrl + Enter), R will try to execute it and, when
ready, will show the results and come back with a new > prompt to wait for new
commands.
If R is still waiting for you to enter more text,
the console will show a + prompt. It means that you haven’t finished entering
a complete command. This is likely because you have not ‘closed’ a parenthesis or
quotation, i.e. you don’t have the same number of left-parentheses as
right-parentheses or the same number of opening and closing quotation marks.
When this happens, and you thought you finished typing your command, click
inside the console window and press Esc; this will cancel the
incomplete command and return you to the > prompt. You can then proofread
the command(s) you entered and correct the error.
Installing additional packages using the packages tab
In addition to the core R installation, there are in excess of 18,000 additional packages which can be used to extend the functionality of R. Many of these have been written by R users and have been made available in central repositories, like the one hosted at CRAN, for anyone to download and install into their own R environment. You should have already installed the packages ‘ggplot2’ and ‘dplyr. If you have not, please do so now using these instructions.
You can see if you have a package installed by looking in the packages tab
(on the lower-right by default). You can also type the command
installed.packages() into the console and examine the output.
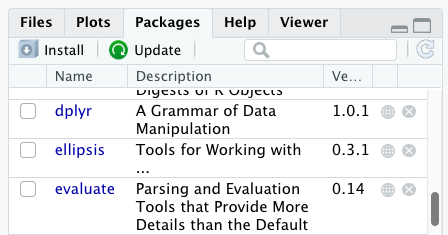
Additional packages can be installed from the ‘packages’ tab. On the packages tab, click the ‘Install’ icon and start typing the name of the package you want in the text box. As you type, packages matching your starting characters will be displayed in a drop-down list so that you can select them.
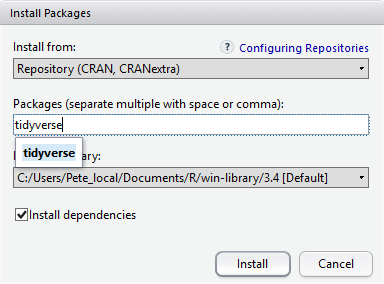
At the bottom of the Install Packages window is a check box to ‘Install’ dependencies. This is ticked by default, which is usually what you want. Packages can (and do) make use of functionality built into other packages, so for the functionality contained in the package you are installing to work properly, there may be other packages which have to be installed with them. The ‘Install dependencies’ option makes sure that this happens.
Exercise
Use both the Console and the Packages tab to confirm that you have the tidyverse installed.
Solution
Scroll through packages tab down to ‘tidyverse’. You can also type a few characters into the searchbox. The ‘tidyverse’ package is really a package of packages, including ‘ggplot2’ and ‘dplyr’, both of which require other packages to run correctly. All of these packages will be installed automatically. Depending on what packages have previously been installed in your R environment, the install of ‘tidyverse’ could be very quick or could take several minutes. As the install proceeds, messages relating to its progress will be written to the console. You will be able to see all of the packages which are actually being installed.
Because the install process accesses the CRAN repository, you will need an Internet connection to install packages.
It is also possible to install packages from other repositories, as well as Github or the local file system, but we won’t be looking at these options in this lesson.
Installing additional packages using R code
If you were watching the console window when you started the install of ‘tidyverse’, you may have noticed that the line
install.packages("tidyverse")
was written to the console before the start of the installation messages.
You could also have installed the tidyverse packages by running this command directly at the R terminal.
We are going to use the library danstat. Please install by running this command:
install.packages("danstat")
If that fails… try this:
install.packages("remotes")
library(remotes)
remotes:install_github("cran/danstat")
Key Points
Use RStudio to write and run R programs.
Use
install.packages()to install packages (libraries).
Introduction to R
Overview
Teaching: 50 min
Exercises: 30 minQuestions
What data types are available in R?
What is an object?
How can values be initially assigned to variables of different data types?
What arithmetic and logical operators can be used?
How can subsets be extracted from vectors?
How does R treat missing values?
How can we deal with missing values in R?
Objectives
A quick recap of the following concepts:
Define the following terms as they relate to R: object, assign, call, function, arguments, options.
Assign values to objects in R.
Learn how to name objects.
Use comments to inform script.
Solve simple arithmetic operations in R.
Call functions and use arguments to change their default options.
Inspect the content of vectors and manipulate their content.
Subset and extract values from vectors.
Analyze vectors with missing data.
A very short refresher on R
You can get output from R simply by typing math in the console:
3 + 5
[1] 8
12 / 7
[1] 1.714286
We can assign values to variables:
area_hectares <- 1.0
<- is the assignment operator. It assigns values on the right to objects on
the left. So, after executing x <- 3, the value of x is 3. The arrow can
be read as 3 goes into x.
Now that R has area_hectares in memory, we can do arithmetic with it. For
instance, we may want to convert this area into acres (area in acres is 2.47 times the area in hectares):
2.47 * area_hectares
[1] 2.47
We can also change an object’s value by assigning it a new one:
area_hectares <- 2.5
2.47 * area_hectares
[1] 6.175
Comments
All programming languages allow the programmer to include comments in their code. To do this in R we use the # character.
Anything to the right of the # sign and up to the end of the line is treated as a comment and is ignored by R. You can start lines with comments
or include them after any code on the line.
area_hectares <- 1.0 # land area in hectares
area_acres <- area_hectares * 2.47 # convert to acres
area_acres # print land area in acres.
[1] 2.47
Functions and their arguments
Functions are “canned scripts” that automate more complicated sets of commands
including operations assignments, etc. Many functions are predefined, or can be
made available by importing R packages (more on that later). A function
usually gets one or more inputs called arguments. Functions often (but not
always) return a value. A typical example would be the function sqrt(). The
input (the argument) must be a number, and the return value (in fact, the
output) is the square root of that number. Executing a function (‘running it’)
is called calling the function. An example of a function call is:
b <- sqrt(a)
Let’s try a function that can take multiple arguments: round().
round(3.14159)
[1] 3
Here, we’ve called round() with just one argument, 3.14159, and it has
returned the value 3.
We can get information on how a function works, with the help function:
?round
We see that if we want a different number of digits, we can
type digits=2 or however many we want.
round(3.14159, digits = 2)
[1] 3.14
Vectors and data types
A vector is the most common and basic data type in R, and is pretty much
the workhorse of R. A vector is composed by a series of values, which can be
either numbers or characters. We can assign a series of values to a vector using
the c() function. For example we can create a vector of the number of household
members for the households we’ve interviewed and assign
it to a new object hh_members:
hh_members <- c(3, 7, 10, 6)
hh_members
[1] 3 7 10 6
A vector can also contain characters. For example, we can have
a vector of the building material used to construct our
interview respondents’ walls (respondent_wall_type):
respondent_wall_type <- c("muddaub", "burntbricks", "sunbricks")
respondent_wall_type
[1] "muddaub" "burntbricks" "sunbricks"
The quotes around “muddaub”, etc. are essential here. Without the quotes R
will assume there are objects called muddaub, burntbricks and sunbricks. As these objects
don’t exist in R’s memory, there will be an error message.
There are many functions that allow you to inspect the content of a
vector. length() tells you how many elements are in a particular vector:
length(hh_members)
[1] 4
length(respondent_wall_type)
[1] 3
An important feature of a vector, is that all of the elements are the same type of data.
The function class() indicates the class (the type of element) of an object:
class(hh_members)
[1] "numeric"
class(respondent_wall_type)
[1] "character"
The function str() provides an overview of the structure of an object and its
elements. It is a useful function when working with large and complex
objects:
str(hh_members)
num [1:4] 3 7 10 6
str(respondent_wall_type)
chr [1:3] "muddaub" "burntbricks" "sunbricks"
You can use the c() function to add other elements to your vector:
possessions <- c("bicycle", "radio", "television")
possessions <- c("car", possessions) # add to the beginning of the vector
possessions
[1] "car" "bicycle" "radio" "television"
In the first line, we take the original vector possessions,
add the value "mobile_phone" to the end of it, and save the result back into
possessions. Then we add the value "car" to the beginning, again saving the result
back into possessions.
We can do this over and over again to grow a vector, or assemble a dataset. As we program, this may be useful to add results that we are collecting or calculating.
Vectors are one of the many data structures that R uses. Other important
ones are lists (list), matrices (matrix), data frames (data.frame),
factors (factor) and arrays (array).
Subsetting vectors
If we want to extract one or several values from a vector, we must provide one or several indices in square brackets. For instance:
respondent_wall_type <- c("muddaub", "burntbricks", "sunbricks")
respondent_wall_type[2]
[1] "burntbricks"
respondent_wall_type[c(3, 2)]
[1] "sunbricks" "burntbricks"
We can also repeat the indices to create an object with more elements than the original one:
more_respondent_wall_type <- respondent_wall_type[c(1, 2, 3, 2, 1, 3)]
more_respondent_wall_type
[1] "muddaub" "burntbricks" "sunbricks" "burntbricks" "muddaub"
[6] "sunbricks"
R indices start at 1. Programming languages like Fortran, MATLAB, Julia, and R start counting at 1, because that’s what human beings typically do. Languages in the C family (including C++, Java, Perl, and Python) count from 0 because that’s simpler for computers to do.
Conditional subsetting
Another common way of subsetting is by using a logical vector. TRUE will
select the element with the same index, while FALSE will not:
hh_members <- c(3, 7, 10, 6)
hh_members[c(TRUE, FALSE, TRUE, TRUE)]
[1] 3 10 6
Typically, these logical vectors are not typed by hand, but are the output of other functions or logical tests. For instance, if you wanted to select only the values above 5:
hh_members > 5 # will return logicals with TRUE for the indices that meet the condition
[1] FALSE TRUE TRUE TRUE
## so we can use this to select only the values above 5
hh_members[hh_members > 5]
[1] 7 10 6
You can combine multiple tests using & (both conditions are true, AND) or |
(at least one of the conditions is true, OR):
hh_members[hh_members < 4 | hh_members > 7]
[1] 3 10
hh_members[hh_members >= 4 & hh_members <= 7]
[1] 7 6
Here, < stands for “less than”, > for “greater than”, >= for “greater than
or equal to”, and == for “equal to”. The double equal sign == is a test for
numerical equality between the left and right hand sides, and should not be
confused with the single = sign, which performs variable assignment (similar
to <-).
A common task is to search for certain strings in a vector. One could use the
“or” operator | to test for equality to multiple values, but this can quickly
become tedious.
possessions <- c("car", "bicycle", "radio", "television", "mobile_phone")
possessions[possessions == "car" | possessions == "bicycle"] # returns both car and bicycle
[1] "car" "bicycle"
The function %in% allows you to test if any of the elements of a search vector
(on the left hand side) are found in the target vector (on the right hand side):
possessions %in% c("car", "bicycle")
[1] TRUE TRUE FALSE FALSE FALSE
Note that the output is the same length as the search vector on the left hand
side, because %in% checks whether each element of the search vector is found
somewhere in the target vector. Thus, you can use %in% to select the elements
in the search vector that appear in your target vector:
possessions %in% c("car", "bicycle", "motorcycle", "truck", "boat", "bus")
[1] TRUE TRUE FALSE FALSE FALSE
possessions[possessions %in% c("car", "bicycle", "motorcycle", "truck", "boat", "bus")]
[1] "car" "bicycle"
Missing data
As R was designed to analyze datasets, it includes the concept of missing data
(which is uncommon in other programming languages). Missing data are represented
in vectors as NA.
When doing operations on numbers, most functions will return NA if the data
you are working with include missing values. This feature
makes it harder to overlook the cases where you are dealing with missing data.
You can add the argument na.rm=TRUE to calculate the result while ignoring
the missing values.
rooms <- c(2, 1, 1, NA, 7)
mean(rooms)
[1] NA
max(rooms)
[1] NA
mean(rooms, na.rm = TRUE)
[1] 2.75
max(rooms, na.rm = TRUE)
[1] 7
If your data include missing values, you may want to become familiar with the
functions is.na(), na.omit(), and complete.cases(). See below for
examples.
Recall that you can use the typeof() function to find the type of your atomic vector.
Key Points
Starting with Data
Overview
Teaching: 50 min
Exercises: 30 minQuestions
What else have we forgotten about R?
What is a data.frame?
How can I read a complete csv file into R?
How can I get basic summary information about my dataset?
How can I change the way R treats strings in my dataset?
Why would I want strings to be treated differently?
How are dates represented in R and how can I change the format?
Objectives
Describe what a data frame is.
Load external data from a .csv file into a data frame.
Summarize the contents of a data frame.
Subset and extract values from data frames.
Describe the difference between a factor and a string.
Convert between strings and factors.
Reorder and rename factors.
Change how character strings are handled in a data frame.
Examine and change date formats.
What are data frames and tibbles?
Data frames are the de facto data structure for tabular data in R, and what
we use for data processing, statistics, and plotting.
Data frames can be created by hand, but most commonly they are generated by the
functions read_csv() or read_table(); in other words, when importing
spreadsheets from your hard drive (or the web). We will now demonstrate how to
import tabular data using read_csv().
Importing data
You are going load the data in R’s memory using the function read_csv()
from the readr package, which is part of the tidyverse; learn
more about the tidyverse collection of packages
here.
readr gets installed as part as the tidyverse installation.
When you load the tidyverse (library(tidyverse)), the core packages
(the packages used in most data analyses) get loaded, including readr.
library(tidyverse)
interviews <- read_csv("../data/SAFI_clean.csv", na = "NULL")
The statement in the code above creates a data frame but doesn’t output
any data because, as you might recall, assignments (<-) don’t display
anything. (Note, however, that read_csv may show informational
text about the data frame that is created.) If we want to check that our data
has been loaded, we can see the contents of the data frame by typing its name:
interviews in the console.
interviews
## Try also
## view(interviews)
## head(interviews)
# A tibble: 131 × 14
key_ID village interview_date no_membrs years_liv respondent_wall… rooms
<dbl> <chr> <dttm> <dbl> <dbl> <chr> <dbl>
1 1 God 2016-11-17 00:00:00 3 4 muddaub 1
2 1 God 2016-11-17 00:00:00 7 9 muddaub 1
3 3 God 2016-11-17 00:00:00 10 15 burntbricks 1
4 4 God 2016-11-17 00:00:00 7 6 burntbricks 1
5 5 God 2016-11-17 00:00:00 7 40 burntbricks 1
6 6 God 2016-11-17 00:00:00 3 3 muddaub 1
7 7 God 2016-11-17 00:00:00 6 38 muddaub 1
8 8 Chirod… 2016-11-16 00:00:00 12 70 burntbricks 3
9 9 Chirod… 2016-11-16 00:00:00 8 6 burntbricks 1
10 10 Chirod… 2016-12-16 00:00:00 12 23 burntbricks 5
# … with 121 more rows, and 7 more variables: memb_assoc <chr>,
# affect_conflicts <chr>, liv_count <dbl>, items_owned <chr>, no_meals <dbl>,
# months_lack_food <chr>, instanceID <chr>
Note
read_csv()assumes that fields are delimited by commas. However, in several countries, the comma is used as a decimal separator and the semicolon (;) is used as a field delimiter. If you want to read in this type of files in R, you can use theread_csv2function. It behaves exactly likeread_csvbut uses different parameters for the decimal and the field separators. If you are working with another format, they can be both specified by the user. Check out the help forread_csv()by typing?read_csvto learn more. There is also theread_tsv()for tab-separated data files, andread_delim()allows you to specify more details about the structure of your file.
Note that read_csv() actually loads the data as a tibble.
A tibble is an extension of R data frames used by the tidyverse. When
the data is read using read_csv(), it is stored in an object of class
tbl_df, tbl, and data.frame. You can see the class of an object with
class(interviews)
[1] "spec_tbl_df" "tbl_df" "tbl" "data.frame"
As a tibble, the type of data included in each column is listed in an
abbreviated fashion below the column names. For instance, here key_ID is a
column of floating point numbers (abbreviated <dbl> for the word ‘double’),
village is a column of characters (<chr>) and the interview_date is a
column in the “date and time” format (<dkttm>).
Inspecting data frames
Size:
dim(interviews)- returns a vector with the number of rows as the first element, and the number of columns as the second element (the dimensions of the object)nrow(interviews)- returns the number of rowsncol(interviews)- returns the number of columns
Content:
head(interviews)- shows the first 6 rowstail(interviews)- shows the last 6 rows
Names:
names(interviews)- returns the column names (synonym ofcolnames()fordata.frameobjects)
Summary:
str(interviews)- structure of the object and information about the class, length and content of each columnsummary(interviews)- summary statistics for each columnglimpse(interviews)- returns the number of columns and rows of the tibble, the names and class of each column, and previews as many values will fit on the screen. Unlike the other inspecting functions listed above,glimpse()is not a “base R” function so you need to have thedplyrortibblepackages loaded to be able to execute it.
Note: most of these functions are “generic.” They can be used on other types of objects besides data frames or tibbles.
Indexing and subsetting data frames
Our interviews data frame has rows and columns (it has 2 dimensions).
In practice, we may not need the entire data frame; for instance, we may only
be interested in a subset of the observations (the rows) or a particular set
of variables (the columns). If we want to
extract some specific data from it, we need to specify the “coordinates” we
want from it. Row numbers come first, followed by column numbers.
Tip
Indexing a
tibblewith[always results in atibble. However, note this is not true in general for data frames, so be careful! Different ways of specifying these coordinates can lead to results with different classes. This is covered in the Software Carpentry lesson R for Reproducible Scientific Analysis.
## first element in the first column of the tibble
interviews[1, 1]
# A tibble: 1 × 1
key_ID
<dbl>
1 1
## first element in the 6th column of the tibble
interviews[1, 6]
# A tibble: 1 × 1
respondent_wall_type
<chr>
1 muddaub
## first column of the tibble (as a vector)
interviews[[1]]
[1] 1 1 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
[19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
[37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 21 54
[55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 127
[73] 133 152 153 155 178 177 180 181 182 186 187 195 196 197 198 201 202 72
[91] 73 76 83 85 89 101 103 102 78 80 104 105 106 109 110 113 118 125
[109] 119 115 108 116 117 144 143 150 159 160 165 166 167 174 175 189 191 192
[127] 126 193 194 199 200
## first column of the tibble
interviews[1]
# A tibble: 131 × 1
key_ID
<dbl>
1 1
2 1
3 3
4 4
5 5
6 6
7 7
8 8
9 9
10 10
# … with 121 more rows
## first three elements in the 7th column of the tibble
interviews[1:3, 7]
# A tibble: 3 × 1
rooms
<dbl>
1 1
2 1
3 1
## the 3rd row of the tibble
interviews[3, ]
# A tibble: 1 × 14
key_ID village interview_date no_membrs years_liv respondent_wall_… rooms
<dbl> <chr> <dttm> <dbl> <dbl> <chr> <dbl>
1 3 God 2016-11-17 00:00:00 10 15 burntbricks 1
# … with 7 more variables: memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>
## equivalent to head_interviews <- head(interviews)
head_interviews <- interviews[1:6, ]
: is a special function that creates numeric vectors of integers in increasing
or decreasing order, test 1:10 and 10:1 for instance.
You can also exclude certain indices of a data frame using the “-” sign:
interviews[, -1] # The whole tibble, except the first column
# A tibble: 131 × 13
village interview_date no_membrs years_liv respondent_wall_type rooms
<chr> <dttm> <dbl> <dbl> <chr> <dbl>
1 God 2016-11-17 00:00:00 3 4 muddaub 1
2 God 2016-11-17 00:00:00 7 9 muddaub 1
3 God 2016-11-17 00:00:00 10 15 burntbricks 1
4 God 2016-11-17 00:00:00 7 6 burntbricks 1
5 God 2016-11-17 00:00:00 7 40 burntbricks 1
6 God 2016-11-17 00:00:00 3 3 muddaub 1
7 God 2016-11-17 00:00:00 6 38 muddaub 1
8 Chirodzo 2016-11-16 00:00:00 12 70 burntbricks 3
9 Chirodzo 2016-11-16 00:00:00 8 6 burntbricks 1
10 Chirodzo 2016-12-16 00:00:00 12 23 burntbricks 5
# … with 121 more rows, and 7 more variables: memb_assoc <chr>,
# affect_conflicts <chr>, liv_count <dbl>, items_owned <chr>, no_meals <dbl>,
# months_lack_food <chr>, instanceID <chr>
interviews[-c(7:131), ] # Equivalent to head(interviews)
# A tibble: 6 × 14
key_ID village interview_date no_membrs years_liv respondent_wall_… rooms
<dbl> <chr> <dttm> <dbl> <dbl> <chr> <dbl>
1 1 God 2016-11-17 00:00:00 3 4 muddaub 1
2 1 God 2016-11-17 00:00:00 7 9 muddaub 1
3 3 God 2016-11-17 00:00:00 10 15 burntbricks 1
4 4 God 2016-11-17 00:00:00 7 6 burntbricks 1
5 5 God 2016-11-17 00:00:00 7 40 burntbricks 1
6 6 God 2016-11-17 00:00:00 3 3 muddaub 1
# … with 7 more variables: memb_assoc <chr>, affect_conflicts <chr>,
# liv_count <dbl>, items_owned <chr>, no_meals <dbl>, months_lack_food <chr>,
# instanceID <chr>
tibbles can be subset by calling indices (as shown previously), but also by
calling their column names directly:
interviews["village"] # Result is a tibble
interviews[, "village"] # Result is a tibble
interviews[["village"]] # Result is a vector
interviews$village # Result is a vector
In RStudio, you can use the autocompletion feature to get the full and correct names of the columns.
Factors
R has a special data class, called factor, to deal with categorical data that you may encounter when creating plots or doing statistical analyses. Factors are very useful and actually contribute to making R particularly well suited to working with data. So we are going to spend a little time introducing them.
Factors represent categorical data. They are stored as integers associated with
labels and they can be ordered (ordinal) or unordered (nominal). Factors
create a structured relation between the different levels (values) of a
categorical variable, such as days of the week or responses to a question in
a survey. This can make it easier to see how one element relates to the
other elements in a column. While factors look (and often behave) like
character vectors, they are actually treated as integer vectors by R. So
you need to be very careful when treating them as strings.
Once created, factors can only contain a pre-defined set of values, known as levels. By default, R always sorts levels in alphabetical order. For instance, if you have a factor with 2 levels:
respondent_floor_type <- factor(c("earth", "cement", "cement", "earth"))
R will assign 1 to the level "cement" and 2 to the level "earth"
(because c comes before e, even though the first element in this vector is
"earth"). You can see this by using the function levels() and you can find
the number of levels using nlevels():
levels(respondent_floor_type)
[1] "cement" "earth"
nlevels(respondent_floor_type)
[1] 2
Sometimes, the order of the factors does not matter. Other times you might want
to specify the order because it is meaningful (e.g., “low”, “medium”, “high”).
It may improve your visualization, or it may be required by a particular type of
analysis. Here, one way to reorder our levels in the respondent_floor_type
vector would be:
respondent_floor_type # current order
[1] earth cement cement earth
Levels: cement earth
respondent_floor_type <- factor(respondent_floor_type,
levels = c("earth", "cement"))
respondent_floor_type # after re-ordering
[1] earth cement cement earth
Levels: earth cement
In R’s memory, these factors are represented by integers (1, 2), but are more
informative than integers because factors are self describing: "cement",
"earth" is more descriptive than 1, and 2. Which one is “earth”? You
wouldn’t be able to tell just from the integer data. Factors, on the other hand,
have this information built in. It is particularly helpful when there are many
levels. It also makes renaming levels easier. Let’s say we made a mistake and
need to recode “cement” to “brick”.
levels(respondent_floor_type)
[1] "earth" "cement"
levels(respondent_floor_type)[2] <- "brick"
levels(respondent_floor_type)
[1] "earth" "brick"
respondent_floor_type
[1] earth brick brick earth
Levels: earth brick
So far, your factor is unordered, like a nominal variable. R does not know the
difference between a nominal and an ordinal variable. You make your factor an
ordered factor by using the ordered=TRUE option inside your factor function.
Note how the reported levels changed from the unordered factor above to the
ordered version below. Ordered levels use the less than sign < to denote
level ranking.
respondent_floor_type_ordered <- factor(respondent_floor_type,
ordered = TRUE)
respondent_floor_type_ordered # after setting as ordered factor
[1] earth brick brick earth
Levels: earth < brick
Converting factors
If you need to convert a factor to a character vector, you use
as.character(x).
as.character(respondent_floor_type)
[1] "earth" "brick" "brick" "earth"
Converting factors where the levels appear as numbers (such as concentration
levels, or years) to a numeric vector is a little trickier. The as.numeric()
function returns the index values of the factor, not its levels, so it will
result in an entirely new (and unwanted in this case) set of numbers.
One method to avoid this is to convert factors to characters, and then to
numbers. Another method is to use the levels() function. Compare:
year_fct <- factor(c(1990, 1983, 1977, 1998, 1990))
as.numeric(year_fct) # Wrong! And there is no warning...
[1] 3 2 1 4 3
as.numeric(as.character(year_fct)) # Works...
[1] 1990 1983 1977 1998 1990
as.numeric(levels(year_fct))[year_fct] # The recommended way.
[1] 1990 1983 1977 1998 1990
Notice that in the recommended levels() approach, three important steps occur:
- We obtain all the factor levels using
levels(year_fct) - We convert these levels to numeric values using
as.numeric(levels(year_fct)) - We then access these numeric values using the underlying integers of the
vector
year_fctinside the square brackets
Renaming factors
When your data is stored as a factor, you can use the plot() function to get a
quick glance at the number of observations represented by each factor level.
Let’s extract the memb_assoc column from our data frame, convert it into a
factor, and use it to look at the number of interview respondents who were or
were not members of an irrigation association:
## create a vector from the data frame column "memb_assoc"
memb_assoc <- interviews$memb_assoc
## convert it into a factor
memb_assoc <- as.factor(memb_assoc)
## let's see what it looks like
memb_assoc
[1] <NA> yes <NA> <NA> <NA> <NA> no yes no no <NA> yes no <NA> yes
[16] <NA> <NA> <NA> <NA> <NA> no <NA> <NA> no no no <NA> no yes <NA>
[31] <NA> yes no yes yes yes <NA> yes <NA> yes <NA> no no <NA> no
[46] no yes <NA> <NA> yes <NA> no yes no <NA> yes no no <NA> no
[61] yes <NA> <NA> <NA> no yes no no no no yes <NA> no yes <NA>
[76] <NA> yes no no yes no no yes no yes no no <NA> yes yes
[91] yes yes yes no no no no yes no no yes yes no <NA> no
[106] no <NA> no no <NA> no <NA> <NA> no no no no yes no no
[121] no no no no no no no no no yes <NA>
Levels: no yes
## bar plot of the number of interview respondents who were
## members of irrigation association:
plot(memb_assoc)
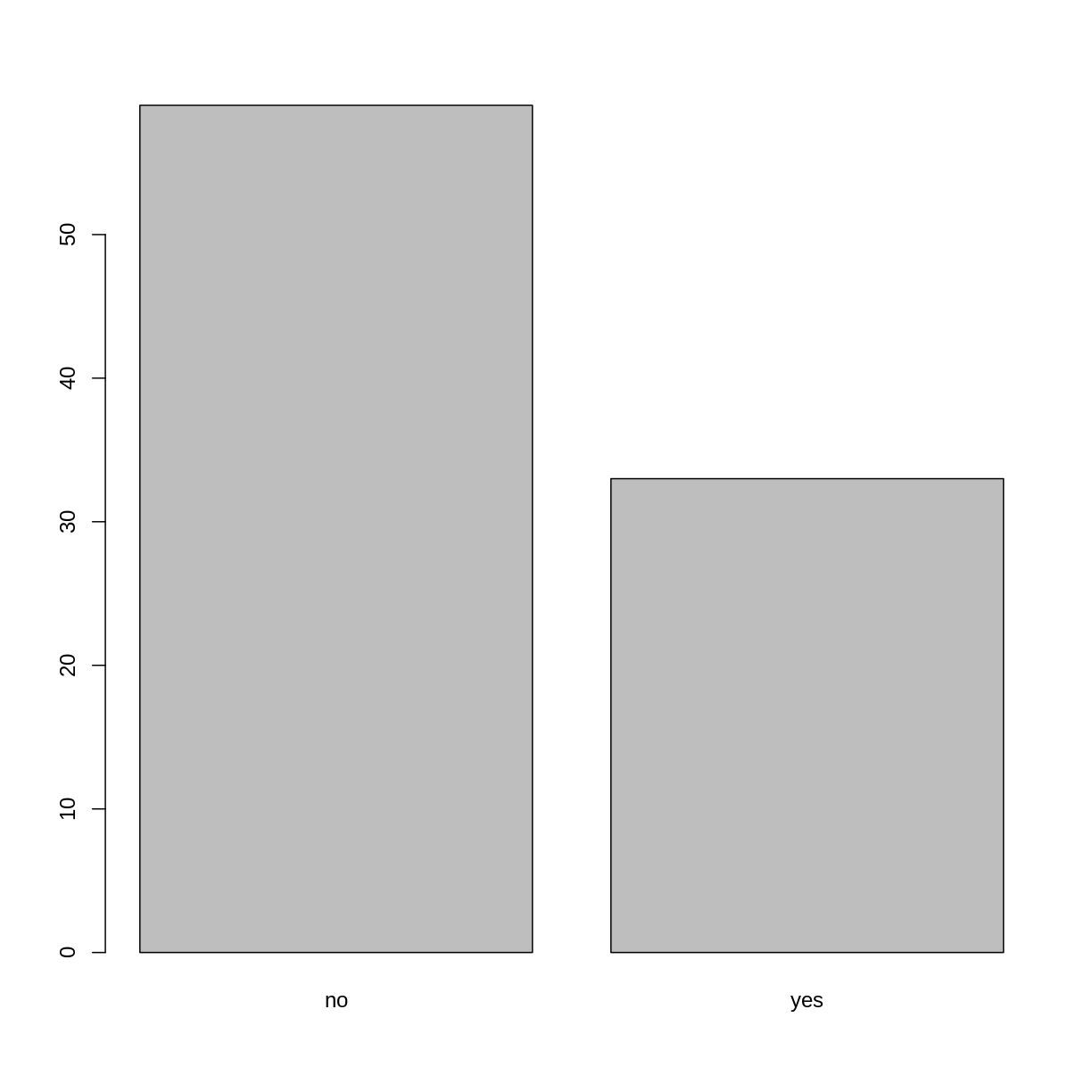
Looking at the plot compared to the output of the vector, we can see that in addition to “no”s and “yes”s, there are some respondents for which the information about whether they were part of an irrigation association hasn’t been recorded, and encoded as missing data. They do not appear on the plot. Let’s encode them differently so they can counted and visualized in our plot.
## Let's recreate the vector from the data frame column "memb_assoc"
memb_assoc <- interviews$memb_assoc
## replace the missing data with "undetermined"
memb_assoc[is.na(memb_assoc)] <- "undetermined"
## convert it into a factor
memb_assoc <- as.factor(memb_assoc)
## let's see what it looks like
memb_assoc
[1] undetermined yes undetermined undetermined undetermined
[6] undetermined no yes no no
[11] undetermined yes no undetermined yes
[16] undetermined undetermined undetermined undetermined undetermined
[21] no undetermined undetermined no no
[26] no undetermined no yes undetermined
[31] undetermined yes no yes yes
[36] yes undetermined yes undetermined yes
[41] undetermined no no undetermined no
[46] no yes undetermined undetermined yes
[51] undetermined no yes no undetermined
[56] yes no no undetermined no
[61] yes undetermined undetermined undetermined no
[66] yes no no no no
[71] yes undetermined no yes undetermined
[76] undetermined yes no no yes
[81] no no yes no yes
[86] no no undetermined yes yes
[91] yes yes yes no no
[96] no no yes no no
[101] yes yes no undetermined no
[106] no undetermined no no undetermined
[111] no undetermined undetermined no no
[116] no no yes no no
[121] no no no no no
[126] no no no no yes
[131] undetermined
Levels: no undetermined yes
## bar plot of the number of interview respondents who were
## members of irrigation association:
plot(memb_assoc)
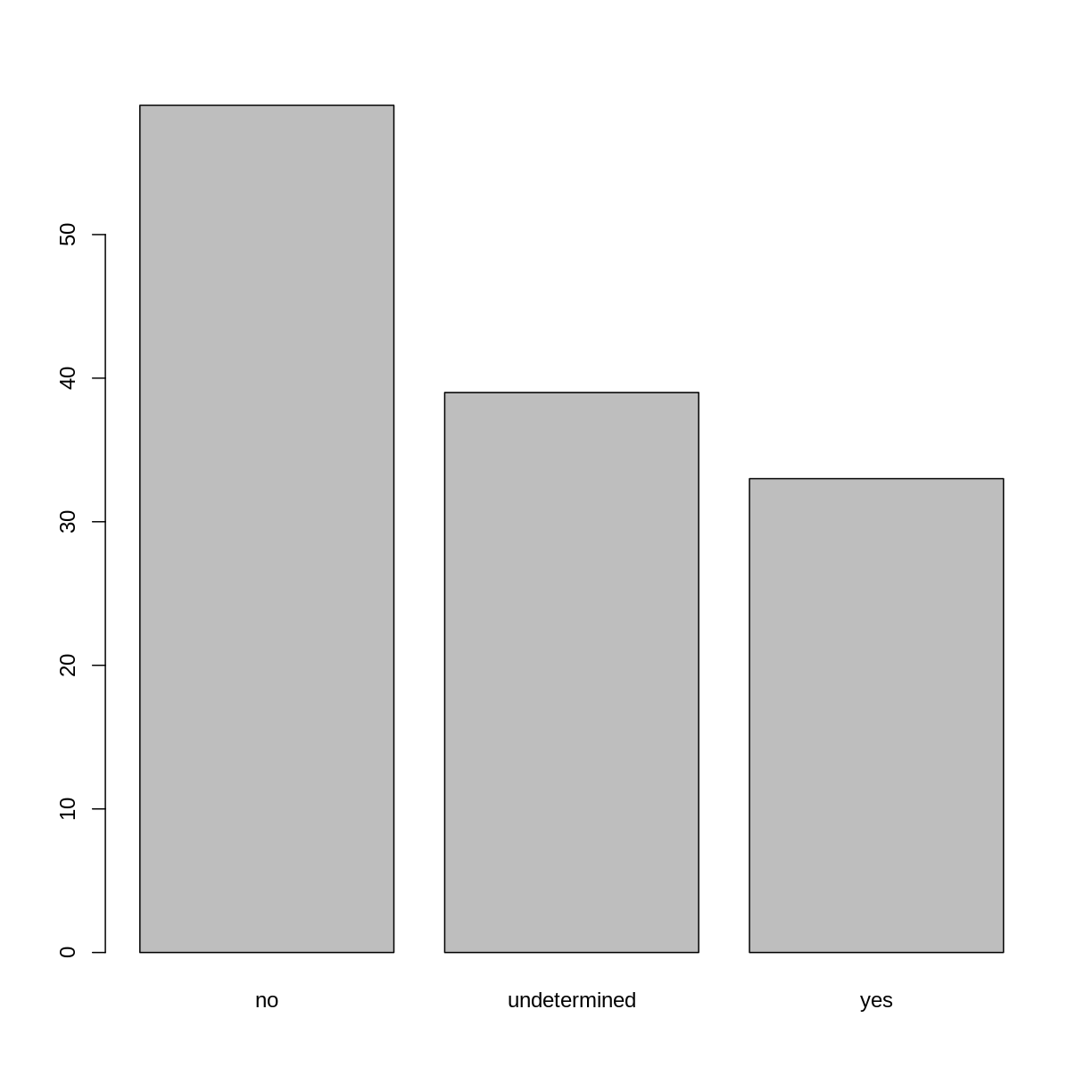
Formatting Dates
One of the most common issues that new (and experienced!) R users have is
converting date and time information into a variable that is appropriate and
usable during analyses. As a reminder from earlier in this lesson, the best
practice for dealing with date data is to ensure that each component of your
date is stored as a separate variable. In our dataset, we have a
column interview_date which contains information about the
year, month, and day that the interview was conducted. Let’s
convert those dates into three separate columns.
str(interviews)
We are going to use the package lubridate, which is included in the
tidyverse installation but not loaded by default, so we have to load
it explicitly with library(lubridate).
Start by loading the required package:
library(lubridate)
The lubridate function ymd() takes a vector representing year, month, and day,
and converts it to a Date vector. Date is a class of data recognized by R as
being a date and can be manipulated as such. The argument that the function
requires is flexible, but, as a best practice, is a character vector formatted
as “YYYY-MM-DD”.
Let’s extract our interview_date column and inspect the structure:
dates <- interviews$interview_date
str(dates)
POSIXct[1:131], format: "2016-11-17" "2016-11-17" "2016-11-17" "2016-11-17" "2016-11-17" ...
When we imported the data in R, read_csv() recognized that this column
contained date information. We can now use the day(), month() and year()
functions to extract this information from the date, and create new columns in
our data frame to store it:
interviews$day <- day(dates)
interviews$month <- month(dates)
interviews$year <- year(dates)
interviews
# A tibble: 131 × 17
key_ID village interview_date no_membrs years_liv respondent_wall… rooms
<dbl> <chr> <dttm> <dbl> <dbl> <chr> <dbl>
1 1 God 2016-11-17 00:00:00 3 4 muddaub 1
2 1 God 2016-11-17 00:00:00 7 9 muddaub 1
3 3 God 2016-11-17 00:00:00 10 15 burntbricks 1
4 4 God 2016-11-17 00:00:00 7 6 burntbricks 1
5 5 God 2016-11-17 00:00:00 7 40 burntbricks 1
6 6 God 2016-11-17 00:00:00 3 3 muddaub 1
7 7 God 2016-11-17 00:00:00 6 38 muddaub 1
8 8 Chirod… 2016-11-16 00:00:00 12 70 burntbricks 3
9 9 Chirod… 2016-11-16 00:00:00 8 6 burntbricks 1
10 10 Chirod… 2016-12-16 00:00:00 12 23 burntbricks 5
# … with 121 more rows, and 10 more variables: memb_assoc <chr>,
# affect_conflicts <chr>, liv_count <dbl>, items_owned <chr>, no_meals <dbl>,
# months_lack_food <chr>, instanceID <chr>, day <int>, month <dbl>,
# year <dbl>
Notice the three new columns at the end of our data frame.
In our example above, the interview_date column was read in correctly as a
Date variable but generally that is not the case. Date columns are often read
in as character variables and one can use the as_date() function to convert
them to the appropriate Date/POSIXctformat.
Let’s say we have a vector of dates in character format:
char_dates <- c("7/31/2012", "8/9/2014", "4/30/2016")
str(char_dates)
chr [1:3] "7/31/2012" "8/9/2014" "4/30/2016"
We can convert this vector to dates as :
as_date(char_dates, format = "%m/%d/%Y")
[1] "2012-07-31" "2014-08-09" "2016-04-30"
Argument format tells the function the order to parse the characters and
identify the month, day and year. The format above is the equivalent of
mm/dd/yyyy. A wrong format can lead to parsing errors or incorrect results.
For example, observe what happens when we use a lower case y instead of upper case Y for the year.
as_date(char_dates, format = "%m/%d/%y")
[1] "2020-07-31" "2020-08-09" "2020-04-30"
Here, the %y part of the format stands for a two-digit year instead of a
four-digit year, and this leads to parsing errors.
Or in the following example, observe what happens when the month and day elements of the format are switched.
as_date(char_dates, format = "%d/%m/%y")
[1] NA "2020-09-08" NA
Since there is no month numbered 30 or 31, the first and third dates cannot be parsed.
We can also use functions ymd(), mdy() or dmy() to convert character
variables to date.
mdy(char_dates)
[1] "2012-07-31" "2014-08-09" "2016-04-30"
Wrangling data with dplyr
dplyr is a package that makes wrangling data easier.
We wrangle data when we select, filter and summarise data.
The pipe construct makes it easy to string together different manipulations of the data:
data %>% filter(some logical test on a column)
We select a set of columns by using the select function:
interviews %>% select(village, memb_assoc)
# A tibble: 131 × 2
village memb_assoc
<chr> <chr>
1 God <NA>
2 God yes
3 God <NA>
4 God <NA>
5 God <NA>
6 God <NA>
7 God no
8 Chirodzo yes
9 Chirodzo no
10 Chirodzo no
# … with 121 more rows
We select a set of rows by using the filter function:
interviews %>% filter(village == "Chirodzo")
# A tibble: 39 × 17
key_ID village interview_date no_membrs years_liv respondent_wall… rooms
<dbl> <chr> <dttm> <dbl> <dbl> <chr> <dbl>
1 8 Chirod… 2016-11-16 00:00:00 12 70 burntbricks 3
2 9 Chirod… 2016-11-16 00:00:00 8 6 burntbricks 1
3 10 Chirod… 2016-12-16 00:00:00 12 23 burntbricks 5
4 34 Chirod… 2016-11-17 00:00:00 8 18 burntbricks 3
5 35 Chirod… 2016-11-17 00:00:00 5 45 muddaub 1
6 36 Chirod… 2016-11-17 00:00:00 6 23 sunbricks 1
7 37 Chirod… 2016-11-17 00:00:00 3 8 burntbricks 1
8 43 Chirod… 2016-11-17 00:00:00 7 29 muddaub 1
9 44 Chirod… 2016-11-17 00:00:00 2 6 muddaub 1
10 45 Chirod… 2016-11-17 00:00:00 9 7 muddaub 1
# … with 29 more rows, and 10 more variables: memb_assoc <chr>,
# affect_conflicts <chr>, liv_count <dbl>, items_owned <chr>, no_meals <dbl>,
# months_lack_food <chr>, instanceID <chr>, day <int>, month <dbl>,
# year <dbl>
We make a new column using the mutate function:
interviews %>% mutate(new_column_name = no_membrs * 10)
# A tibble: 131 × 18
key_ID village interview_date no_membrs years_liv respondent_wall… rooms
<dbl> <chr> <dttm> <dbl> <dbl> <chr> <dbl>
1 1 God 2016-11-17 00:00:00 3 4 muddaub 1
2 1 God 2016-11-17 00:00:00 7 9 muddaub 1
3 3 God 2016-11-17 00:00:00 10 15 burntbricks 1
4 4 God 2016-11-17 00:00:00 7 6 burntbricks 1
5 5 God 2016-11-17 00:00:00 7 40 burntbricks 1
6 6 God 2016-11-17 00:00:00 3 3 muddaub 1
7 7 God 2016-11-17 00:00:00 6 38 muddaub 1
8 8 Chirod… 2016-11-16 00:00:00 12 70 burntbricks 3
9 9 Chirod… 2016-11-16 00:00:00 8 6 burntbricks 1
10 10 Chirod… 2016-12-16 00:00:00 12 23 burntbricks 5
# … with 121 more rows, and 11 more variables: memb_assoc <chr>,
# affect_conflicts <chr>, liv_count <dbl>, items_owned <chr>, no_meals <dbl>,
# months_lack_food <chr>, instanceID <chr>, day <int>, month <dbl>,
# year <dbl>, new_column_name <dbl>
We calculate summary statistics by using the summarize function:
interviews %>% summarise(avg_membrs = mean(no_membrs))
# A tibble: 1 × 1
avg_membrs
<dbl>
1 7.19
Summary statistics are normally combined with the function group_by:
interviews %>% group_by(village) %>%
summarise(avg_membrs = mean(no_membrs))
# A tibble: 3 × 2
village avg_membrs
<chr> <dbl>
1 Chirodzo 7.08
2 God 6.86
3 Ruaca 7.57
Key Points
Use read_csv to read tabular data in R.
Use factors to represent categorical data in R.
What is an API?
Overview
Teaching: 30 min
Exercises: 15 minQuestions
What is an API?
Objectives
Understand what an API do
Connect to Statistics Denmark, and extract data
Create a list of lists to control the variables to be extracted
Please note: These pages are autogenerated. Some of the API-calls may fail during that process. We are figuring out what to do about it, but please excuse us for any red errors on the pages for the time being.
What is an API?
An API is an Application Programming Interface. It is a way of making applications, in our case an R-script, able to communicate with another application, here the Statistics Denmark databases.
Talking about APIs, we talk about several different things. It can be quite confusing, but dont worry!
What we want to be able to do, is to let our own application, our R-script, send a command to a remote application, the databases of Statistics Denmark, in order to retrieve specific data.
This is equivalent to requesting a page from a webserver.
The HTTP protocol can be visualized like this:
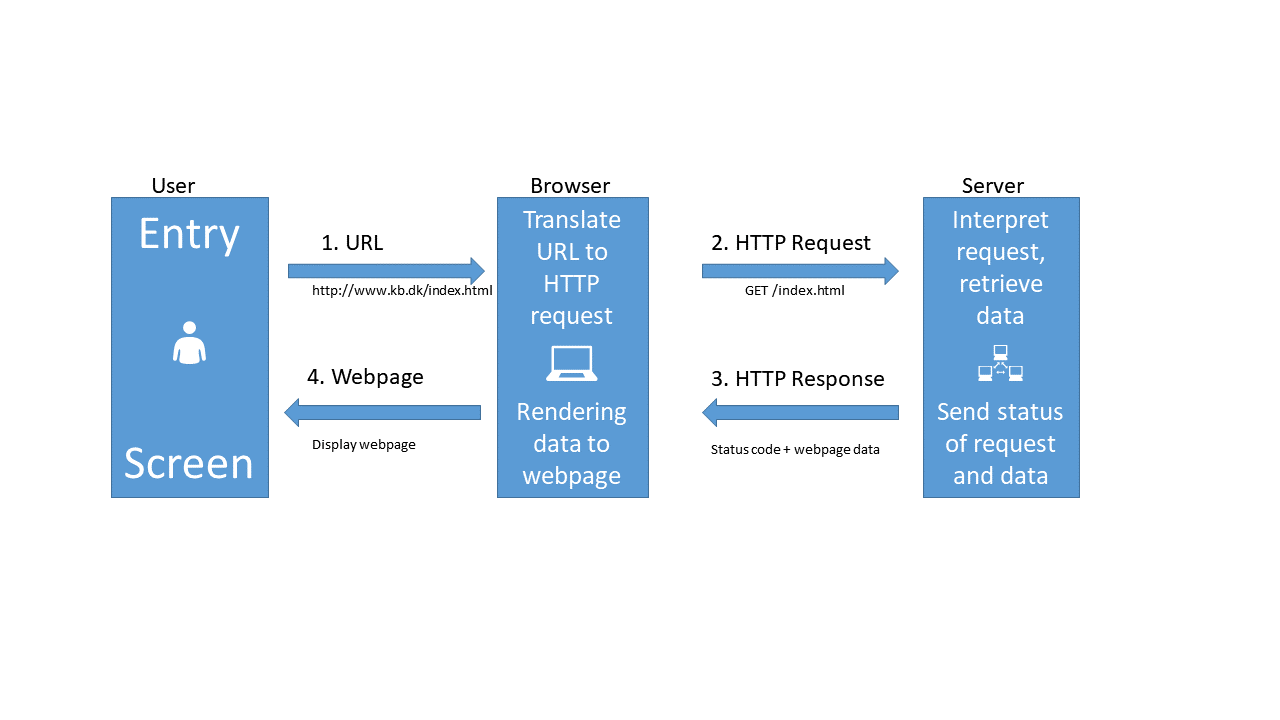
-
When we type in an URL in our browser, it translates that URL to a HTTP-request.
-
The browser sends that HTTP-request to a webserver. The request contains information about the page we need, but in the “header” of the request, there is a lot of other information. The version of browser we are using and cookies to just mention two.
-
The webserver interpret the request, and retrieves the data.
-
After that, the webserver sends both the status of the request (hopefully 200 - which is short for “everything is OK”), and the data.
-
The browser receives the data, and displays it as a webpage.
When we are working with APIs we cut out the user. We have a script that needs some data. We write code that defines, and then send a request til a server, specifying which data we need. The server extracts the needed data, and returns it to the script.
So - how do we do that?
Looking closer at the illustration above, we can see that we send a GET-request to the server. But we are not only asking for at simple page, we need to specify some more information. And then we have to use a slightly different request to the server, a POST-request.
With a POST-request we can control what data is send along with the request, and the data returned by the server depends on what data we send.
We are going to write a POST-request (with a little help from R), to retrieve data from Statistics Denmark.
But before we can do that, we need to know how the SD-API expects to receive data.
Hopefully we can get that by reading the documentation. We can find that here:
https://www.dst.dk/en/Statistik/brug-statistikken/muligheder-i-statistikbanken/api
That was confusing!
Three main things:
Statistics Denmark provides four “functions”, or “endpoints”:

The first is the “web”-site we have to send requests to if we want information on the subjects in Statistics Denmark.
In the second we get information about which tables are available for a given subject.
The third will provide metadata on a table.
When we finally need the data, we will visit the last endpoint.
Let us send a request to subjects.
The endpoint was
endpoint <- "http://api.statbank.dk/v1/subjects"
We will now need to construct a named list for the content of the body that we send along with our request.
This is a new datastructure that we have not encountered before.
Vectors are annoying because they can only contain one datatype. And dataframes must be rectangular.
A list allows us to store basically anything. The reason that we dont use them generally is that they are a bit more difficult to work with.
our_body <- list(lang = "en", recursive = FALSE,
includeTables = FALSE, subjects = NULL)
This list contains four elements, with names. The first, lang, contains a character vector (lenght 1), containing “en”, the language that we want Statistics Denmark to use when returning data.
recursive and includeTables are logical values, both false. And subjects is a special value, NULL. This is not a missing value, there simply isn’t anything there. But this nothing does have a name.
Now we have the two things we need, an endpoint to send a request, and a body containg what we want returned.
Let us try it:
result <- httr::POST(endpoint, body=our_body, encode = "json")
We ask to get the result in json, a speciel datastructure that is able to contain almost anything.
Let us look at the result:
result
Response [https://api.statbank.dk/v1/subjects]
Date: 2022-04-01 10:54
Status: 200
Content-Type: text/json; charset=utf-8
Size: 884 B
Both informative. And utterly useless. The informative information is that our request succeeded (cave - it might not succeed on this webpage). We can see that in the status. 200 is an internet code for success.
Let us get the content of the result, which is what we actually want:
httr::content(result)
[1] "[{\"id\":\"1\",\"description\":\"People\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]},{\"id\":\"2\",\"description\":\"Labour and income\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]},{\"id\":\"3\",\"description\":\"Economy\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]},{\"id\":\"4\",\"description\":\"Social conditions\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]},{\"id\":\"5\",\"description\":\"Education and research\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]},{\"id\":\"6\",\"description\":\"Business\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]},{\"id\":\"7\",\"description\":\"Transport\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]},{\"id\":\"8\",\"description\":\"Culture and leisure\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]},{\"id\":\"9\",\"description\":\"Environment and energy\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]},{\"id\":\"19\",\"description\":\"Other\",\"active\":true,\"hasSubjects\":true,\"subjects\":[]}]"
More informative, but not really easy to read.
The library jsonlite has a function that converts this to something readable:
jsonlite::fromJSON(httr::content(result))
id description active hasSubjects subjects
1 1 People TRUE TRUE NULL
2 2 Labour and income TRUE TRUE NULL
3 3 Economy TRUE TRUE NULL
4 4 Social conditions TRUE TRUE NULL
5 5 Education and research TRUE TRUE NULL
6 6 Business TRUE TRUE NULL
7 7 Transport TRUE TRUE NULL
8 8 Culture and leisure TRUE TRUE NULL
9 9 Environment and energy TRUE TRUE NULL
10 19 Other TRUE TRUE NULL
A nice dataframe with the ten major subjects in the databases of Statistics Denmark.
Subject 1 contains information about populations and elections.
There are sub-subjects under that. We now modify our body that we send with the request, to return information about the first subject.
We need to make sure that the number of the subject, 1 is intepreted as it is. This is a little bit of mysterious handwaving - we simply put the 1 inside the function I() and stuff works.
our_body <- list(lang = "en", recursive = F,
includeTables = F, subjects = I(1))
Note that it is important that we tell the POST function that the body is the body:
data <- httr::POST(endpoint, body=our_body, encode = "json") %>%
httr::content() %>%
jsonlite::fromJSON()
data
id description active hasSubjects
1 1 People TRUE TRUE
subjects
1 3401, 3407, 3410, 3415, 3412, 3411, 3428, 3409, Population, Households, families and children, Migration, Housing, Health, Democracy, National church, Names, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE
We now get at data frame containg a dataframe. We pick that out:
data$subjects
[[1]]
id description active hasSubjects subjects
1 3401 Population TRUE TRUE NULL
2 3407 Households, families and children TRUE TRUE NULL
3 3410 Migration TRUE TRUE NULL
4 3415 Housing TRUE TRUE NULL
5 3412 Health TRUE TRUE NULL
6 3411 Democracy TRUE TRUE NULL
7 3428 National church TRUE TRUE NULL
8 3409 Names TRUE TRUE NULL
This was why the dollar-notation for subsetting dataframes is important.
These are the sub-subjects of subject 1.
Let us look closer at 3401, Population.
Again, we modify the call we send to the endpoint:
our_body <- list(lang = "en", recursive = F,
includeTables = F, subjects = I(3401))
data <- httr::POST(endpoint, body=our_body, encode = "json") %>%
httr::content() %>%
jsonlite::fromJSON()
data
id description active hasSubjects
1 3401 Population TRUE TRUE
subjects
1 20021, 20024, 20022, 20019, 20017, 20018, 20014, 20015, Population figures, Immigrants and their descendants, Population projections, Adoptions, Births, Fertility, Deaths, Life expectancy, TRUE, TRUE, TRUE, FALSE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE
We delve deeper into it:
data$subjects
[[1]]
id description active hasSubjects subjects
1 20021 Population figures TRUE FALSE NULL
2 20024 Immigrants and their descendants TRUE FALSE NULL
3 20022 Population projections TRUE FALSE NULL
4 20019 Adoptions FALSE FALSE NULL
5 20017 Births TRUE FALSE NULL
6 20018 Fertility TRUE FALSE NULL
7 20014 Deaths TRUE FALSE NULL
8 20015 Life expectancy TRUE FALSE NULL
And now we are at the bottom. 20021 Population figures does not have any sub-sub-subjects.
Next, let us take a look at the tables contained under subject 20021.
We need the next endpoint, which provides information about tables under a subject:
endpoint <- "http://api.statbank.dk/v1/tables"
our_body <- list(lang = "en", subjects = I(20021))
data <- httr::POST(endpoint, body=our_body, encode = "json") %>%
httr::content() %>%
jsonlite::fromJSON()
data
id text
1 FOLK1A Population at the first day of the quarter
2 FOLK1AM Population at the first day of the month
3 FOLK3 Population 1. January
4 FOLK3FOD Population 1. January
5 BEF5 Population 1. January
6 FT Population figures from the censuses
7 BY1 Population 1. January
8 BY2 Population 1. January
9 BY3 Population 1. January
10 KM1 Population at the first day of the quarter
11 SOGN1 Population 1. January
12 SOGN10 Population 1. January
13 BEF4 Population 1. January
14 BEF5F People born in Faroe Islands and living in Denmark 1. January
15 BEF5G People born in Greenland and living in Denmark 1. January
16 BEV22 Summary vital statistics (provisional data)
17 BEV107 Summary vital statistics
18 KMSTA003 Summary vital statistics
19 GALDER Average age
20 KMGALDER Average age
21 HISB3 Summary vital statistics
unit updated firstPeriod latestPeriod active
1 Number 2022-02-11T08:00:00 2008Q1 2022Q1 TRUE
2 Number 2022-03-07T08:00:00 2021M10 2022M02 TRUE
3 Number 2022-02-11T08:00:00 2008 2022 TRUE
4 Number 2022-03-18T08:00:00 2008 2022 TRUE
5 Number 2022-02-11T08:00:00 1990 2022 TRUE
6 Number 2022-02-11T08:00:00 1769 2022 TRUE
7 Number 2021-04-29T08:00:00 2010 2021 TRUE
8 Number 2021-04-29T08:00:00 2010 2021 TRUE
9 - 2021-04-29T08:00:00 2017 2021 TRUE
10 Number 2022-02-17T08:00:00 2007Q1 2022Q1 TRUE
11 Number 2022-02-17T08:00:00 2010 2022 TRUE
12 Number 2021-09-22T08:00:00 1925 2021 TRUE
13 Number 2021-03-31T08:00:00 1901 2021 TRUE
14 Number 2022-02-11T08:00:00 2008 2022 TRUE
15 Number 2022-02-11T08:00:00 2008 2022 TRUE
16 Number 2022-02-11T08:00:00 2007Q2 2021Q4 TRUE
17 Number 2022-02-11T08:00:00 2006 2021 TRUE
18 Number 2022-02-17T08:00:00 2015 2021 TRUE
19 Average 2022-02-11T08:00:00 2005 2022 TRUE
20 Average 2022-02-17T08:00:00 2007 2022 TRUE
21 Number 2022-02-11T08:00:00 1901 2022 TRUE
variables
1 region, sex, age, marital status, time
2 region, sex, age, time
3 day of birth, birth month, year of birth, time
4 day of birth, birth month, country of birth, time
5 sex, age, country of birth, time
6 national part, time
7 urban and rural areas, age, sex, time
8 municipality, city size, age, sex, time
9 urban and rural areas, population, area and population density, time
10 parish, member of the National Church, time
11 parish, sex, age, time
12 parish, time
13 islands, time
14 sex, age, parents place of birth, time
15 sex, age, parents place of birth, time
16 region, type of movement, sex, time
17 region, type of movement, sex, time
18 parish, movements, time
19 municipality, sex, time
20 parish, sex, time
21 type of movement, time
There are 21 tables under this subject. Let us see what information we can get about table “FOLK1A”:
We now need the third endpoint:
endpoint <- "http://api.statbank.dk/v1/tableinfo"
our_body <- list(lang = "en", table = "FOLK1A")
data <- httr::POST(endpoint, body=our_body, encode = "json") %>%
httr::content() %>%
jsonlite::fromJSON()
data
$id
[1] "FOLK1A"
$text
[1] "Population at the first day of the quarter"
$description
[1] "Population at the first day of the quarter by region, sex, age, marital status and time"
$unit
[1] "Number"
$suppressedDataValue
[1] "0"
$updated
[1] "2022-02-11T08:00:00"
$active
[1] TRUE
$contacts
name phone mail
1 Dorthe Larsen +4539173307 dla@dst.dk
$documentation
$documentation$id
[1] "4a12721d-a8b0-4bde-82d7-1d1c6f319de3"
$documentation$url
[1] "https://www.dst.dk/documentationofstatistics/4a12721d-a8b0-4bde-82d7-1d1c6f319de3"
$footnote
NULL
$variables
id text elimination time map
1 OMRÅDE region TRUE FALSE denmark_municipality_07
2 KØN sex TRUE FALSE <NA>
3 ALDER age TRUE FALSE <NA>
4 CIVILSTAND marital status TRUE FALSE <NA>
5 Tid time FALSE TRUE <NA>
values
1 000, 084, 101, 147, 155, 185, 165, 151, 153, 157, 159, 161, 163, 167, 169, 183, 173, 175, 187, 201, 240, 210, 250, 190, 270, 260, 217, 219, 223, 230, 400, 411, 085, 253, 259, 350, 265, 269, 320, 376, 316, 326, 360, 370, 306, 329, 330, 340, 336, 390, 083, 420, 430, 440, 482, 410, 480, 450, 461, 479, 492, 530, 561, 563, 607, 510, 621, 540, 550, 573, 575, 630, 580, 082, 710, 766, 615, 707, 727, 730, 741, 740, 746, 706, 751, 657, 661, 756, 665, 760, 779, 671, 791, 081, 810, 813, 860, 849, 825, 846, 773, 840, 787, 820, 851, All Denmark, Region Hovedstaden, Copenhagen, Frederiksberg, Dragør, Tårnby, Albertslund, Ballerup, Brøndby, Gentofte, Gladsaxe, Glostrup, Herlev, Hvidovre, Høje-Taastrup, Ishøj, Lyngby-Taarbæk, Rødovre, Vallensbæk, Allerød, Egedal, Fredensborg, Frederikssund, Furesø, Gribskov, Halsnæs, Helsingør, Hillerød, Hørsholm, Rudersdal, Bornholm, Christiansø, Region Sjælland, Greve, Køge, Lejre, Roskilde, Solrød, Faxe, Guldborgsund, Holbæk, Kalundborg, Lolland, Næstved, Odsherred, Ringsted, Slagelse, Sorø, Stevns, Vordingborg, Region Syddanmark, Assens, Faaborg-Midtfyn, Kerteminde, Langeland, Middelfart, Nordfyns, Nyborg, Odense, Svendborg, Ærø, Billund, Esbjerg, Fanø, Fredericia, Haderslev, Kolding, Sønderborg, Tønder, Varde, Vejen, Vejle, Aabenraa, Region Midtjylland, Favrskov, Hedensted, Horsens, Norddjurs, Odder, Randers, Samsø, Silkeborg, Skanderborg, Syddjurs, Aarhus, Herning, Holstebro, Ikast-Brande, Lemvig, Ringkøbing-Skjern, Skive, Struer, Viborg, Region Nordjylland, Brønderslev, Frederikshavn, Hjørring, Jammerbugt, Læsø, Mariagerfjord, Morsø, Rebild, Thisted, Vesthimmerlands, Aalborg
2 TOT, 1, 2, Total, Men, Women
3 IALT, 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, Total, 0 years, 1 year, 2 years, 3 years, 4 years, 5 years, 6 years, 7 years, 8 years, 9 years, 10 years, 11 years, 12 years, 13 years, 14 years, 15 years, 16 years, 17 years, 18 years, 19 years, 20 years, 21 years, 22 years, 23 years, 24 years, 25 years, 26 years, 27 years, 28 years, 29 years, 30 years, 31 years, 32 years, 33 years, 34 years, 35 years, 36 years, 37 years, 38 years, 39 years, 40 years, 41 years, 42 years, 43 years, 44 years, 45 years, 46 years, 47 years, 48 years, 49 years, 50 years, 51 years, 52 years, 53 years, 54 years, 55 years, 56 years, 57 years, 58 years, 59 years, 60 years, 61 years, 62 years, 63 years, 64 years, 65 years, 66 years, 67 years, 68 years, 69 years, 70 years, 71 years, 72 years, 73 years, 74 years, 75 years, 76 years, 77 years, 78 years, 79 years, 80 years, 81 years, 82 years, 83 years, 84 years, 85 years, 86 years, 87 years, 88 years, 89 years, 90 years, 91 years, 92 years, 93 years, 94 years, 95 years, 96 years, 97 years, 98 years, 99 years, 100 years, 101 years, 102 years, 103 years, 104 years, 105 years, 106 years, 107 years, 108 years, 109 years, 110 years, 111 years, 112 years, 113 years, 114 years, 115 years, 116 years, 117 years, 118 years, 119 years, 120 years, 121 years, 122 years, 123 years, 124 years, 125 years
4 TOT, U, G, E, F, Total, Never married, Married/separated, Widowed, Divorced
5 2008K1, 2008K2, 2008K3, 2008K4, 2009K1, 2009K2, 2009K3, 2009K4, 2010K1, 2010K2, 2010K3, 2010K4, 2011K1, 2011K2, 2011K3, 2011K4, 2012K1, 2012K2, 2012K3, 2012K4, 2013K1, 2013K2, 2013K3, 2013K4, 2014K1, 2014K2, 2014K3, 2014K4, 2015K1, 2015K2, 2015K3, 2015K4, 2016K1, 2016K2, 2016K3, 2016K4, 2017K1, 2017K2, 2017K3, 2017K4, 2018K1, 2018K2, 2018K3, 2018K4, 2019K1, 2019K2, 2019K3, 2019K4, 2020K1, 2020K2, 2020K3, 2020K4, 2021K1, 2021K2, 2021K3, 2021K4, 2022K1, 2008Q1, 2008Q2, 2008Q3, 2008Q4, 2009Q1, 2009Q2, 2009Q3, 2009Q4, 2010Q1, 2010Q2, 2010Q3, 2010Q4, 2011Q1, 2011Q2, 2011Q3, 2011Q4, 2012Q1, 2012Q2, 2012Q3, 2012Q4, 2013Q1, 2013Q2, 2013Q3, 2013Q4, 2014Q1, 2014Q2, 2014Q3, 2014Q4, 2015Q1, 2015Q2, 2015Q3, 2015Q4, 2016Q1, 2016Q2, 2016Q3, 2016Q4, 2017Q1, 2017Q2, 2017Q3, 2017Q4, 2018Q1, 2018Q2, 2018Q3, 2018Q4, 2019Q1, 2019Q2, 2019Q3, 2019Q4, 2020Q1, 2020Q2, 2020Q3, 2020Q4, 2021Q1, 2021Q2, 2021Q3, 2021Q4, 2022Q1
This is a bit more complicated. We are told that:
-
there are five columns in this table.
-
They each have an id
-
And a descriptive text
-
Elimination means that the API will attempt to eliminate the variables we have not chosen values for when data is returned. This makes sense when we get to point 7.
-
time - only one of the variables contain information about a point in time.
-
One of the variables can be mapped to - well a map
-
The final column provides information about which values are stored in the variable. There are 105 different regions in Denmark. And if we do not choose a specific region - the API will attempt to eliminate this facetting, and return data for all of Denmark.
These data provides useful information for constructing the final call to the API in order to get the data.
We will now need the final endpoint:
endpoint <- "http://api.statbank.dk/v1/data"
And we will need to specify which information, from which table, we want data in the body of the request. That is a bit more complicated. We need to make a list of lists!
variables <- list(list(code = "OMRÅDE", values = I("*")),
list(code = "CIVILSTAND", values = I(c("U", "G", "E", "F"))),
list(code = "Tid", values = I("*"))
)
our_body <- list(table = "FOLK1A", lang = "en", format = "CSV", variables = variables)
The final endpoint is:
endpoint <- "https://api.statbank.dk/v1/data"
And the call:
data <- httr::POST(endpoint, body=our_body, encode = "json")
The data is returned as csv - we defined that in “our_body”, so we now need to extract it a bit differently:
data <- data %>%
httr::content(type = "text") %>%
read_csv2()
ℹ Using "','" as decimal and "'.'" as grouping mark. Use `read_delim()` for more control.
No encoding supplied: defaulting to UTF-8.
Rows: 23940 Columns: 4
── Column specification ────────────────────────────────────────────────────────
Delimiter: ";"
chr (3): OMRÅDE, CIVILSTAND, TID
dbl (1): INDHOLD
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
data
# A tibble: 23,940 × 4
OMRÅDE CIVILSTAND TID INDHOLD
<chr> <chr> <chr> <dbl>
1 All Denmark Never married 2008Q1 2552700
2 All Denmark Never married 2008Q2 2563134
3 All Denmark Never married 2008Q3 2564705
4 All Denmark Never married 2008Q4 2568255
5 All Denmark Never married 2009Q1 2575185
6 All Denmark Never married 2009Q2 2584993
7 All Denmark Never married 2009Q3 2584560
8 All Denmark Never married 2009Q4 2588198
9 All Denmark Never married 2010Q1 2593172
10 All Denmark Never married 2010Q2 2604129
# … with 23,930 more rows
Voila! We have a dataframe with information about how many persons in Denmark were married (or not) at different points in time.
That was a bit complicated. There are easier ways to do it.
We will look at that shortly. So why do it this way? These techniques are the same techniques we use when we access an arbitrary other API. The fields, endpoints etc might be different. We might have an added complication of having to login to it. But the techniques can be reused.
Key Points
Getting data from an API is equivalent to requesting a webpage
POST requests to servers put specific demands on how we request data
What about danstat?
Overview
Teaching: 30 min
Exercises: 15 minQuestions
An easier way to access Statistics Denmark
Objectives
Understand what an API do
Connect to Statistics Denmark, and extract data
Create a list of lists to control the variables to be extracted
Please note: These pages are autogenerated. Some of the API-calls may fail during that process. We are figuring out what to do about it, but please excuse us for any red errors on the pages for the time being.
What is an API?
An API is an Application Programming Interface. It is a way of making applications, in our case an R-script, able to communicate with another application, here the Statistics Denmark databases.
Talking about APIs, we talk about several different things. It can be quite confusing, but dont worry!
What we want to be able to do, is to let our own application, our R-script, send a command to a remote application, the databases of Statistics Denmark, in order to retrieve specific data.
An API defines the different commands we can send, and how the data that we get back, is formatted.
Often APIs will require a user account with a login and a password. Statistics Denmark does not.
The standard way to send a command, or a request, to an API is to use the GET (and POST) functions at the core of the internet.
In a certain sense this is what we do when we access a website. We go to www.dr.dk/sporten and get a result, the current webpage at the front of the sports section of Danmarks Radio.
If we instead ask www.dr.dk to return the result of our request for www.dr.dk/nyheder/politik, we will get the current webpage with news on politics.
This is what we do when we access an API. But instead of using our browser, we use the method our browser uses (GET), tells that method that we would like some specified information, and get a result that is not a webpage, but rather a set of data. Hopefully organised in a way that is easy to read.
Writing our own GET-requests to communicate with an API is not simple. Thankfully kind people have written libraries, some in R, that makes accessing specific APIs easier. The one we are going to use here is called “danstat”
The danstat package/library
Before doing anything else, it is useful to take a look at the result:
# A tibble: 6 × 4
IELAND KØN TID INDHOLD
<chr> <chr> <chr> <dbl>
1 Denmark Men 2008Q1 2465810
2 Denmark Men 2008Q2 2466036
3 Denmark Men 2008Q3 2467712
4 Denmark Men 2008Q4 2469977
5 Denmark Men 2009Q1 2470457
6 Denmark Men 2009Q2 2470287
This is from the table “folk1c” from Statistics Denmark.
We get some variables, IELAND, KØN, and TID. And then the content of the table, INDHOLD. Ie the number of men, living in denmark i the first quarter of 2008 in the first line.
How do we get that table?
All tables from Statistics Denmark are organised in a hierarcical tree of subjects.
Let us begin there.
Before using the library, we need to install it:
install.packages("danstat")
Some installations of R may have problems installing it. In that case, try this:
install.packages("remotes")
library(remotes)
remotes:install_github("cran/danstat")
After installation, we load the library using the library function. And then we can access the functions included in the library:
The get_subjects() function sends a request to the Statistics Denmark API, asking for a list of the subjects. The information is returned to our script, and the get_subjects() function presents us with a dataframe containing the information.
library(danstat)
subjects <- get_subjects()
subjects
id description active hasSubjects subjects
1 1 People TRUE TRUE NULL
2 2 Labour and income TRUE TRUE NULL
3 3 Economy TRUE TRUE NULL
4 4 Social conditions TRUE TRUE NULL
5 5 Education and research TRUE TRUE NULL
6 6 Business TRUE TRUE NULL
7 7 Transport TRUE TRUE NULL
8 8 Culture and leisure TRUE TRUE NULL
9 9 Environment and energy TRUE TRUE NULL
10 19 Other TRUE TRUE NULL
We get the 13 major subjects from Statistics Denmark. Each of them have sub-subjects.
If we want to take a closer look at the subdivisions of a given subject, we use the get_subjects() function again, this time specifying which subject we are interested in:
Let us try to get the sub-subjects from the subject 1 - containing information about populations and elections:
sub_subjects <- get_subjects(subjects = 1)
sub_subjects
id description active hasSubjects
1 1 People TRUE TRUE
subjects
1 3401, 3407, 3410, 3415, 3412, 3411, 3428, 3409, Population, Households, families and children, Migration, Housing, Health, Democracy, National church, Names, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE
The result is a bit complicated. The column “subjects” in the resulting dataframe contains another dataframe. We access it like we normally would access a column in a dataframe:
sub_subjects$subjects
[[1]]
id description active hasSubjects subjects
1 3401 Population TRUE TRUE NULL
2 3407 Households, families and children TRUE TRUE NULL
3 3410 Migration TRUE TRUE NULL
4 3415 Housing TRUE TRUE NULL
5 3412 Health TRUE TRUE NULL
6 3411 Democracy TRUE TRUE NULL
7 3428 National church TRUE TRUE NULL
8 3409 Names TRUE TRUE NULL
Those sub-subjects have their own subjects! Lets get to the bottom of this, and use 2401, Population and population projections as an example:
sub_sub_subjects <- get_subjects("3401")
sub_sub_subjects$subjects
[[1]]
id description active hasSubjects subjects
1 20021 Population figures TRUE FALSE NULL
2 20024 Immigrants and their descendants TRUE FALSE NULL
3 20022 Population projections TRUE FALSE NULL
4 20019 Adoptions FALSE FALSE NULL
5 20017 Births TRUE FALSE NULL
6 20018 Fertility TRUE FALSE NULL
7 20014 Deaths TRUE FALSE NULL
8 20015 Life expectancy TRUE FALSE NULL
Now we are at the bottom. We can see in the column “hasSubjects” that there are no sub_sub_sub_subjects.
The hierarchy is: 1 Population and elections | 3401 Population | 20021 Population figures
The final sub_sub_subject contains a number of tables, that actually contains the data we are looking for.
get_subjects is able to retrieve all the sub, sub-sub and sub-sub-sub-jects in one go. The result is a bit confusing and difficult to navigate.
Remember that the initial result was a dataframe containing another dataframe. If we go all the way to the bottom, we will get a dataframe, containing several dataframes, each of those containing several dataframes.
We recommend that you do not try it, but this is how it is done:
lots_of_subjects <- get_subjects(1, recursive = T, include_tables = T)
The “recursive = T” parameter means that get_subjects will retrieve the subjects of the subjects, and then the subjects of those subjects.
Which datatables exists?
But we ended up with a sub_sub_subject,
20021 Population figures
How do we find out which tables exists in this subject?
The get_tables() function returns a dataframe with information about the tables available for a given subject.
tables <- get_tables(subjects="20021")
tables
id text
1 FOLK1A Population at the first day of the quarter
2 FOLK1AM Population at the first day of the month
3 FOLK3 Population 1. January
4 FOLK3FOD Population 1. January
5 BEF5 Population 1. January
6 FT Population figures from the censuses
7 BY1 Population 1. January
8 BY2 Population 1. January
9 BY3 Population 1. January
10 KM1 Population at the first day of the quarter
11 SOGN1 Population 1. January
12 SOGN10 Population 1. January
13 BEF4 Population 1. January
14 BEF5F People born in Faroe Islands and living in Denmark 1. January
15 BEF5G People born in Greenland and living in Denmark 1. January
16 BEV22 Summary vital statistics (provisional data)
17 BEV107 Summary vital statistics
18 KMSTA003 Summary vital statistics
19 GALDER Average age
20 KMGALDER Average age
21 HISB3 Summary vital statistics
unit updated firstPeriod latestPeriod active
1 Number 2022-02-11T08:00:00 2008Q1 2022Q1 TRUE
2 Number 2022-03-07T08:00:00 2021M10 2022M02 TRUE
3 Number 2022-02-11T08:00:00 2008 2022 TRUE
4 Number 2022-03-18T08:00:00 2008 2022 TRUE
5 Number 2022-02-11T08:00:00 1990 2022 TRUE
6 Number 2022-02-11T08:00:00 1769 2022 TRUE
7 Number 2021-04-29T08:00:00 2010 2021 TRUE
8 Number 2021-04-29T08:00:00 2010 2021 TRUE
9 - 2021-04-29T08:00:00 2017 2021 TRUE
10 Number 2022-02-17T08:00:00 2007Q1 2022Q1 TRUE
11 Number 2022-02-17T08:00:00 2010 2022 TRUE
12 Number 2021-09-22T08:00:00 1925 2021 TRUE
13 Number 2021-03-31T08:00:00 1901 2021 TRUE
14 Number 2022-02-11T08:00:00 2008 2022 TRUE
15 Number 2022-02-11T08:00:00 2008 2022 TRUE
16 Number 2022-02-11T08:00:00 2007Q2 2021Q4 TRUE
17 Number 2022-02-11T08:00:00 2006 2021 TRUE
18 Number 2022-02-17T08:00:00 2015 2021 TRUE
19 Average 2022-02-11T08:00:00 2005 2022 TRUE
20 Average 2022-02-17T08:00:00 2007 2022 TRUE
21 Number 2022-02-11T08:00:00 1901 2022 TRUE
variables
1 region, sex, age, marital status, time
2 region, sex, age, time
3 day of birth, birth month, year of birth, time
4 day of birth, birth month, country of birth, time
5 sex, age, country of birth, time
6 national part, time
7 urban and rural areas, age, sex, time
8 municipality, city size, age, sex, time
9 urban and rural areas, population, area and population density, time
10 parish, member of the National Church, time
11 parish, sex, age, time
12 parish, time
13 islands, time
14 sex, age, parents place of birth, time
15 sex, age, parents place of birth, time
16 region, type of movement, sex, time
17 region, type of movement, sex, time
18 parish, movements, time
19 municipality, sex, time
20 parish, sex, time
21 type of movement, time
We get at lot of information here. The id identifies the table, text gives a description of the table that humans can understand. When the table was last updated and the first and last period that the table contains data for.
In the variables column, we get information on what kind of data is stored in the table.
Before we pull out the data, we need to know which variables are available in the table. We do this with this function:
metadata <- get_table_metadata("FOLK1A", variables_only = T)
metadata
id text elimination time map
1 OMRÅDE region TRUE FALSE denmark_municipality_07
2 KØN sex TRUE FALSE <NA>
3 ALDER age TRUE FALSE <NA>
4 CIVILSTAND marital status TRUE FALSE <NA>
5 Tid time FALSE TRUE <NA>
values
1 000, 084, 101, 147, 155, 185, 165, 151, 153, 157, 159, 161, 163, 167, 169, 183, 173, 175, 187, 201, 240, 210, 250, 190, 270, 260, 217, 219, 223, 230, 400, 411, 085, 253, 259, 350, 265, 269, 320, 376, 316, 326, 360, 370, 306, 329, 330, 340, 336, 390, 083, 420, 430, 440, 482, 410, 480, 450, 461, 479, 492, 530, 561, 563, 607, 510, 621, 540, 550, 573, 575, 630, 580, 082, 710, 766, 615, 707, 727, 730, 741, 740, 746, 706, 751, 657, 661, 756, 665, 760, 779, 671, 791, 081, 810, 813, 860, 849, 825, 846, 773, 840, 787, 820, 851, All Denmark, Region Hovedstaden, Copenhagen, Frederiksberg, Dragør, Tårnby, Albertslund, Ballerup, Brøndby, Gentofte, Gladsaxe, Glostrup, Herlev, Hvidovre, Høje-Taastrup, Ishøj, Lyngby-Taarbæk, Rødovre, Vallensbæk, Allerød, Egedal, Fredensborg, Frederikssund, Furesø, Gribskov, Halsnæs, Helsingør, Hillerød, Hørsholm, Rudersdal, Bornholm, Christiansø, Region Sjælland, Greve, Køge, Lejre, Roskilde, Solrød, Faxe, Guldborgsund, Holbæk, Kalundborg, Lolland, Næstved, Odsherred, Ringsted, Slagelse, Sorø, Stevns, Vordingborg, Region Syddanmark, Assens, Faaborg-Midtfyn, Kerteminde, Langeland, Middelfart, Nordfyns, Nyborg, Odense, Svendborg, Ærø, Billund, Esbjerg, Fanø, Fredericia, Haderslev, Kolding, Sønderborg, Tønder, Varde, Vejen, Vejle, Aabenraa, Region Midtjylland, Favrskov, Hedensted, Horsens, Norddjurs, Odder, Randers, Samsø, Silkeborg, Skanderborg, Syddjurs, Aarhus, Herning, Holstebro, Ikast-Brande, Lemvig, Ringkøbing-Skjern, Skive, Struer, Viborg, Region Nordjylland, Brønderslev, Frederikshavn, Hjørring, Jammerbugt, Læsø, Mariagerfjord, Morsø, Rebild, Thisted, Vesthimmerlands, Aalborg
2 TOT, 1, 2, Total, Men, Women
3 IALT, 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, Total, 0 years, 1 year, 2 years, 3 years, 4 years, 5 years, 6 years, 7 years, 8 years, 9 years, 10 years, 11 years, 12 years, 13 years, 14 years, 15 years, 16 years, 17 years, 18 years, 19 years, 20 years, 21 years, 22 years, 23 years, 24 years, 25 years, 26 years, 27 years, 28 years, 29 years, 30 years, 31 years, 32 years, 33 years, 34 years, 35 years, 36 years, 37 years, 38 years, 39 years, 40 years, 41 years, 42 years, 43 years, 44 years, 45 years, 46 years, 47 years, 48 years, 49 years, 50 years, 51 years, 52 years, 53 years, 54 years, 55 years, 56 years, 57 years, 58 years, 59 years, 60 years, 61 years, 62 years, 63 years, 64 years, 65 years, 66 years, 67 years, 68 years, 69 years, 70 years, 71 years, 72 years, 73 years, 74 years, 75 years, 76 years, 77 years, 78 years, 79 years, 80 years, 81 years, 82 years, 83 years, 84 years, 85 years, 86 years, 87 years, 88 years, 89 years, 90 years, 91 years, 92 years, 93 years, 94 years, 95 years, 96 years, 97 years, 98 years, 99 years, 100 years, 101 years, 102 years, 103 years, 104 years, 105 years, 106 years, 107 years, 108 years, 109 years, 110 years, 111 years, 112 years, 113 years, 114 years, 115 years, 116 years, 117 years, 118 years, 119 years, 120 years, 121 years, 122 years, 123 years, 124 years, 125 years
4 TOT, U, G, E, F, Total, Never married, Married/separated, Widowed, Divorced
5 2008K1, 2008K2, 2008K3, 2008K4, 2009K1, 2009K2, 2009K3, 2009K4, 2010K1, 2010K2, 2010K3, 2010K4, 2011K1, 2011K2, 2011K3, 2011K4, 2012K1, 2012K2, 2012K3, 2012K4, 2013K1, 2013K2, 2013K3, 2013K4, 2014K1, 2014K2, 2014K3, 2014K4, 2015K1, 2015K2, 2015K3, 2015K4, 2016K1, 2016K2, 2016K3, 2016K4, 2017K1, 2017K2, 2017K3, 2017K4, 2018K1, 2018K2, 2018K3, 2018K4, 2019K1, 2019K2, 2019K3, 2019K4, 2020K1, 2020K2, 2020K3, 2020K4, 2021K1, 2021K2, 2021K3, 2021K4, 2022K1, 2008Q1, 2008Q2, 2008Q3, 2008Q4, 2009Q1, 2009Q2, 2009Q3, 2009Q4, 2010Q1, 2010Q2, 2010Q3, 2010Q4, 2011Q1, 2011Q2, 2011Q3, 2011Q4, 2012Q1, 2012Q2, 2012Q3, 2012Q4, 2013Q1, 2013Q2, 2013Q3, 2013Q4, 2014Q1, 2014Q2, 2014Q3, 2014Q4, 2015Q1, 2015Q2, 2015Q3, 2015Q4, 2016Q1, 2016Q2, 2016Q3, 2016Q4, 2017Q1, 2017Q2, 2017Q3, 2017Q4, 2018Q1, 2018Q2, 2018Q3, 2018Q4, 2019Q1, 2019Q2, 2019Q3, 2019Q4, 2020Q1, 2020Q2, 2020Q3, 2020Q4, 2021Q1, 2021Q2, 2021Q3, 2021Q4, 2022Q1
There is a lot of other metadata in the tables, including the phone number to the staffmember at Statistics Denmark that is responsible for maintaining the table. We are only interested in the variables, which is why we add the parameter “variables_only = T”.
What kind of values can the individual datapoints take?
metadata %>% slice(4) %>% pull(values)
[[1]]
id text
1 TOT Total
2 U Never married
3 G Married/separated
4 E Widowed
5 F Divorced
We use the slice function from tidyverse to pull out the fourth row of the dataframe, and the pull-function to pull out the values in the values column.
The same trick can be done for the other fields in the table:
metadata %>% slice(1) %>% pull(values) %>% .[[1]] %>% head
id text
1 000 All Denmark
2 084 Region Hovedstaden
3 101 Copenhagen
4 147 Frederiksberg
5 155 Dragør
6 185 Tårnby
Here we see the individual municipalities in Denmark.
Now we are almost ready to pull out the actual data!
But first!
Which variables do we want?
We need to specify which variables we want in our answer. Do we want the total population for all municipalities in Denmark? Or just a few? Do we want the total population, or do we want it broken down by sex.
These variables, and the values of them, need to be specified when we pull the data from Statistics Denmark.
We also need to provide that information in a specific way.
If we want data for all municipalites, we want to pull the variable “OMRÅDE” from the list of variables.
Therefore we need to give the function an argument containing both the information that we want the population data broken down by “OMRÅDE”, and that we want all values of “OMRÅDE”.
Vectors are characterized by only being able to contain one type of data.
When we need to have structures that can contain more than one type of data, we can use the list structure.
Lists allows us to have values, with names (sometime descriptive).
Lists can even contain lists.
And that is what we need here. Let us make our first list:
list(code = "OMRÅDE", values = NA)
$code
[1] "OMRÅDE"
$values
[1] NA
This list have to components. One called “code”, and one called “values”. Code have the content “OMRÅDE”, specifying that we want the variable in the data from Statistics Denmark calld “OMRÅDE”.
“values” has the content “NA”. We use “NA”, when we want to specify that we want all the “OMRÅDE”. If we only wanted a specific municipality, we could instead specify it instead of writing “NA”.
Let us assume that we also want to break down the data based on marriage status.
That information is stored in the variable “CIVILSTAND”.
And above, we saw that we had the following values in that variable:
metadata %>% slice(4) %>% pull(values)
[[1]]
id text
1 TOT Total
2 U Never married
3 G Married/separated
4 E Widowed
5 F Divorced
A value for the total population is probably not that interesting, if we pull all the individual values for “Never married” etc.
We can now make another list:
list(code = "CIVILSTAND", values = c("U", "G", "E", "F"))
$code
[1] "CIVILSTAND"
$values
[1] "U" "G" "E" "F"
Here the “values” part is a vector containing the values we want to pull out for that variable.
It might be interesting to take a look at how the population changes over time.
In that case we need to pull out data from the “Tid” variable.
That would look like this:
list(code = "Tid", values = NA)
$code
[1] "Tid"
$values
[1] NA
If we want to pull data broken down by all three variables, we need to provide a list, containing three lists.
We do that using this code:
variables <- list(list(code = "OMRÅDE", values = NA),
list(code = "CIVILSTAND", values = c("U", "G", "E", "F")),
list(code = "Tid", values = NA)
)
variables
[[1]]
[[1]]$code
[1] "OMRÅDE"
[[1]]$values
[1] NA
[[2]]
[[2]]$code
[1] "CIVILSTAND"
[[2]]$values
[1] "U" "G" "E" "F"
[[3]]
[[3]]$code
[1] "Tid"
[[3]]$values
[1] NA
And now, finally, we are ready to get the data!
data <- get_data(table_id = "FOLK1A", variables = variables)
Rows: 23940 Columns: 4
── Column specification ────────────────────────────────────────────────────────
Delimiter: ";"
chr (3): OMRÅDE, CIVILSTAND, TID
dbl (1): INDHOLD
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
It takes a short moment. But now we have a dataframe containing the data we requested:
head(data)
# A tibble: 6 × 4
OMRÅDE CIVILSTAND TID INDHOLD
<chr> <chr> <chr> <dbl>
1 All Denmark Never married 2008Q1 2552700
2 All Denmark Never married 2008Q2 2563134
3 All Denmark Never married 2008Q3 2564705
4 All Denmark Never married 2008Q4 2568255
5 All Denmark Never married 2009Q1 2575185
6 All Denmark Never married 2009Q2 2584993
This procedure will work for all the tables from Statistics Denmark!
The data is nicely formatted and ready to use. Almost.
Before we do anything else, let us save the data.
write_csv2(data, "../data/SD_data.csv")
Key Points
R Markdown is a useful language for creating reproducible documents combining text and executable R-code.
Time
Overview
Teaching: 50 min
Exercises: 30 minQuestions
How can I select specific rows and/or columns from a dataframe?
Objectives
Describe the purpose of an R package and the
dplyrandtidyrpackages.Select certain columns in a dataframe with the
dplyrfunctionselect.
A relatively short session on time.
“People assume that time is a strict progression from cause to effect, but actually from a non-linear, non-subjective viewpoint, it’s more like a big ball of wibbly-wobbly, timey-wimey stuff.”
Time is not easy to deal with. It is actually really complicated. Here is a rant on how complicated it is…
https://www.youtube.com/watch?v=-5wpm-gesOY
Why?
We just pulled data out giving us the danish population, broken down by marriage status and geographical area. And time.
If the data is not still in memory, we can read it in:
data <- read_csv2("../data/SD_data.csv")
ℹ Using "','" as decimal and "'.'" as grouping mark. Use `read_delim()` for more control.
Rows: 23100 Columns: 4
── Column specification ────────────────────────────────────────────────────────
Delimiter: ";"
chr (3): OMRÅDE, CIVILSTAND, TID
dbl (1): INDHOLD
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
head(data)
# A tibble: 6 × 4
OMRÅDE CIVILSTAND TID INDHOLD
<chr> <chr> <chr> <dbl>
1 All Denmark Never married 2008Q1 2552700
2 All Denmark Never married 2008Q2 2563134
3 All Denmark Never married 2008Q3 2564705
4 All Denmark Never married 2008Q4 2568255
5 All Denmark Never married 2009Q1 2575185
6 All Denmark Never married 2009Q2 2584993
Note that the datatype for “TID” is
A general tool
lubridate is a package written to make working with dates and times easy(er).
It may need to be installed first.
install.packages("lubridate")
After that, we can load it:
library(lubridate)
Attaching package: 'lubridate'
The following objects are masked from 'package:base':
date, intersect, setdiff, union
Lubridate converts a lot of different ways of writing dates to a consistent date-time format.
The most important functions we need to know, are:
- ymd
- hms
- ymd_hms
And variations of these, especially ymd.
ymd(“2021-09-21”) converts the date 2020-09-21 to a date-format that R can understand:
ymd("2021-09-21")
[1] "2021-09-21"
Sometimes we have dates formatted as “21-09-2021”. That is day, month and year in that order.
That can be converted to at standard date-format with the function dmy():
dmy("21-09-2021")
[1] "2021-09-21"
We might even have dates formatted as “2021 21 4”, (year, day month), the function ydm() can handle that.
ydm("2021 21 4")
[1] "2021-04-21"
Time is handled in a similar way, but time is usually not written as creatively as dates:
hm("14:05")
[1] "14H 5M 0S"
hms("14.05.21")
[1] "14H 5M 21S"
Dates and times can be combined, as in: “2021-04-21 14:05:12”:
ymd_hms("2021-04-21 14:05:12")
[1] "2021-04-21 14:05:12 UTC"
Those were the nice dates…
Not so nice date formats - a more specific tool
Statistics Denmark returns a lot of data-series by quarter, or month. And we need to convert it to something we can work with. Without necessarily understanding all the details.
The library tsibble provides functions that can convert “2020Q1”, the first quarter of 2020, into something R can understand as time-value:
We might need to install it first:
install.packages("tsibble")
And then load it:
library(tsibble)
Attaching package: 'tsibble'
The following object is masked from 'package:lubridate':
interval
The following objects are masked from 'package:base':
intersect, setdiff, union
This is a vector containg the 8 quarters of the years 2019 and 2020.
quarters <- c("2019Q1", "2019Q2", "2019Q3", "2019Q4", "2020Q1", "2020Q2", "2020Q3", "2020Q4")
class(quarters)
[1] "character"
It is a character vector, ie strings. If we want to analyse any data associated with these specific quarters, we need to convert them to something R is able to recognize as time.
yearquarter(quarters)
<yearquarter[8]>
[1] "2019 Q1" "2019 Q2" "2019 Q3" "2019 Q4" "2020 Q1" "2020 Q2" "2020 Q3"
[8] "2020 Q4"
# Year starts on: January
We are not going to go into further details on the challenges of working with time-series. The generic lubridate functions and yearquarter() will be enough for our purposes.
Let us finish by converting the “TID” column in our data, to a time-format.
data <- data %>%
mutate(TID = yearquarter(TID))
We mutate the column “TID” into the result of running yearquarter() on the column “TID”. And now we have a data frame that we can do interesting things with.
Now might be a good time to save the data in its new version:
write_csv2(data, "../data/SD_data.csv")
Note that we are using write_csv2() here. We do not have decimalpoints in this data, but other data might have.
Key Points
Use
pivot_longer()to go from wide to long format.
Data Visualisation with ggplot2
Overview
Teaching: 80 min
Exercises: 35 minQuestions
What are the components of a ggplot?
How do I create scatterplots, boxplots, and barplots?
How can I change the aesthetics (ex. colour, transparency) of my plot?
How can I create multiple plots at once?
Objectives
Produce scatter plots, boxplots, and barplots using ggplot.
Set universal plot settings.
Describe what faceting is and apply faceting in ggplot.
Modify the aesthetics of an existing ggplot plot (including axis labels and colour).
Build complex and customized plots from data in a data frame.
Nice data. How does it look?
R has some nice plotting functions build in.
ggplot2 is a package with more, nicer, plotting possibilities.
We start by loading the required package. ggplot2 is also included in the
tidyverse package.
library(tidyverse)
If not still in the workspace, load the data we saved in the previous lesson.
SD_data <- read_csv2("../data/SD_data.csv")
ℹ Using "','" as decimal and "'.'" as grouping mark. Use `read_delim()` for more control.
Rows: 23940 Columns: 4
── Column specification ────────────────────────────────────────────────────────
Delimiter: ";"
chr (3): OMRÅDE, CIVILSTAND, TID
dbl (1): INDHOLD
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
We read in data from a csv-file. That is stored as text, so we need to convert the “TID” column to something that can be understood as time by R:
SD_data <- SD_data %>% mutate(TID = yearquarter(TID))
Plotting with ggplot2
ggplot2 is a plotting package that makes it simple to create complex plots
from data stored in a data frame. It provides a programmatic interface for
specifying what variables to plot, how they are displayed, and general visual
properties. Therefore, we only need minimal changes if the underlying data
change or if we decide to change from a bar plot to a scatterplot. This helps in
creating publication quality plots with minimal amounts of adjustments and
tweaking.
ggplot2 functions work best with data in the ‘long’ format, i.e., a column for every
dimension, and a row for every observation. Well-structured data will save you
lots of time when making figures with ggplot2
ggplot graphics are built step by step by adding new elements. Adding layers in this fashion allows for extensive flexibility and customization of plots.
Each chart built with ggplot2 must include the following
- Data
-
Aesthetic mapping (aes)
- Describes how variables are mapped onto graphical attributes
- Visual attribute of data including x-y axes, color, fill, shape, and alpha
-
Geometric objects (geom)
- Determines how values are rendered graphically, as bars (
geom_bar), scatterplot (geom_point), line (geom_line), etc.
- Determines how values are rendered graphically, as bars (
Thus, the template for graphic in ggplot2 is:
<DATA> %>%
ggplot(aes(<MAPPINGS>)) +
<GEOM_FUNCTION>()
Remember from the last lesson that the pipe operator %>% places the result of the previous line(s) into the first argument of the function. ggplot is a function that expects a data frame to be the first argument. This allows for us to change from specifying the data = argument within the ggplot function and instead pipe the data into the function.
- use the
ggplot()function and bind the plot to a specific data frame.
SD_data %>%
ggplot()
- define a mapping (using the aesthetic (
aes) function), by selecting the variables to be plotted and specifying how to present them in the graph, e.g. as x/y positions or characteristics such as size, shape, color, etc.
SD_data %>%
ggplot(aes(x = TID, y = INDHOLD))
-
add ‘geoms’ – graphical representations of the data in the plot (points, lines, bars).
ggplot2offers many different geoms; we will use some common ones today, including:geom_point()for scatter plots, dot plots, etc.geom_boxplot()for, well, boxplots!geom_line()for trend lines, time series, etc.
To add a geom to the plot use the + operator. Because we have two continuous variables, let’s use geom_point() first:
SD_data %>%
ggplot(aes(x = TID, y = INDHOLD)) +
geom_point()
 What we might note that the fact that we have ALL the municipalites leads to a
LOT of points.
What we might note that the fact that we have ALL the municipalites leads to a
LOT of points.
We could have done that when we extracted the data from Statistics Denmark. Alternatively we can do it now. Let us pull out all the regions.
plot_data <- SD_data %>%
filter(str_detect(OMRÅDE, "Region"))
We use the filter function - we have seen before. And it returns the rows in the data where the expression we write in the paranthesis is true.
From the package “stringr”, included in the tidyverse package, we get the function str_detect().
It detects if the string “Region” is present in the variable OMRÅDE. If it is, “Region” is detected, the expression is true, and filter() leaves the row.
Back to ggplot2
The + in the ggplot2 package is particularly useful because it allows
you to modify existing ggplot objects. This means you can easily set up plot
templates and conveniently explore different types of plots, so the above plot
can also be generated with code like this, similar to the “intermediate steps”
approach in the previous lesson. We are now plotting the plot_data dataframe
instead:
# Assign plot to a variable
data_plot <- plot_data %>%
ggplot(aes(x = TID, y = INDHOLD))
# Draw the plot as a dot plot
data_plot +
geom_point()
 A lot better.
A lot better.
Notes
- Anything you put in the
ggplot()function can be seen by any geom layers that you add (i.e., these are universal plot settings). This includes the x- and y-axis mapping you set up inaes().- You can also specify mappings for a given geom independently of the mapping defined globally in the
ggplot()function.- The
+sign used to add new layers must be placed at the end of the line containing the previous layer. If, instead, the+sign is added at the beginning of the line containing the new layer,ggplot2will not add the new layer and will return an error message.
## This is the correct syntax for adding layers
data_plot +
geom_point()
## This will not add the new layer and will return an error message
data_plot
+ geom_point()
Building your plots iteratively
Building plots with ggplot2 is typically an iterative process. We start by
defining the dataset we’ll use, lay out the axes, and choose a geom:
plot_data %>%
ggplot(aes(x = TID, y = INDHOLD)) +
geom_point()
Then, we start modifying this plot to extract more information from it. We might want to color the points, based on the marriage status.
We place the color argument within the aes() function, because we want to map the values in “CIVILSTAND” to the
plot_data %>%
ggplot(aes(x = TID, y = INDHOLD, color = CIVILSTAND)) +
geom_point()
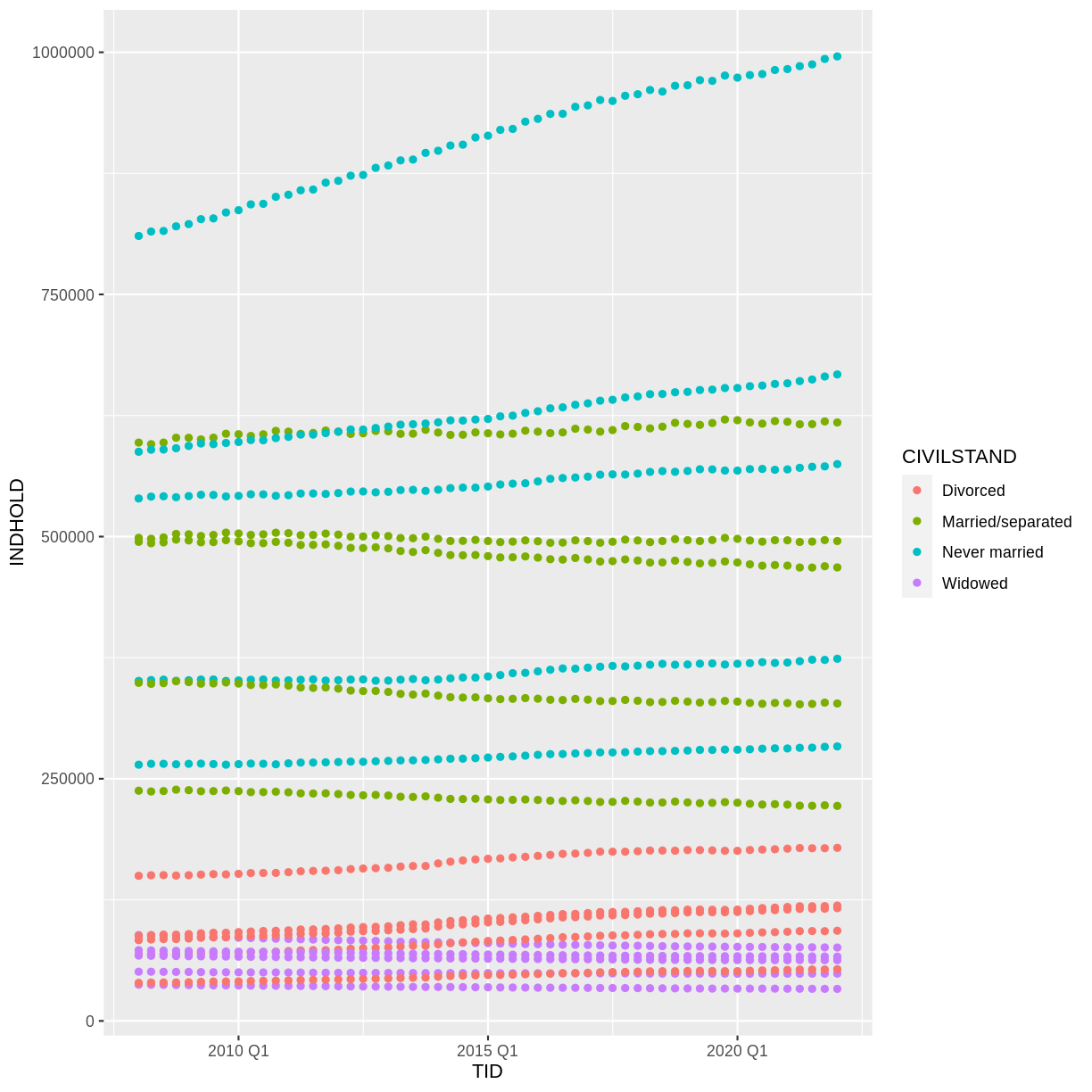
To colour each marriage status in the plot differently, you could use a vector as an input
to the argument color. However, because we are now mapping features of the
data to a colour, instead of setting one colour for all points, the colour of the
points now needs to be set inside a call to the aes function. When we map
a variable in our data to the colour of the points, ggplot2 will provide a
different colour corresponding to the different values of the variable. We will
continue to specify the value of alpha, width, and height
outside of the aes function because we are using the same value for
every point. ggplot2 understands both the Commonwealth English and
American English spellings for colour, i.e., you can use either color
or colour. The plot aboge is an example where we color points by
the CIVILSTAND of the observation.
Faceting
We still have a lot of information Rather than creating a single plot with points for each region, we may want to create multiple plot, where each plot shows the data for a single region.
ggplot2 has a special technique called faceting that allows the
user to split one plot into multiple plots based on a factor included
in the dataset. We will use it to split our plot of CIVILSTAND
against time, by OMRÅDE, so each region has its own panel in a
multi-panel plot:
plot_data %>%
ggplot(aes(x = TID, y = INDHOLD, color = CIVILSTAND)) +
geom_point() +
facet_wrap(~OMRÅDE)
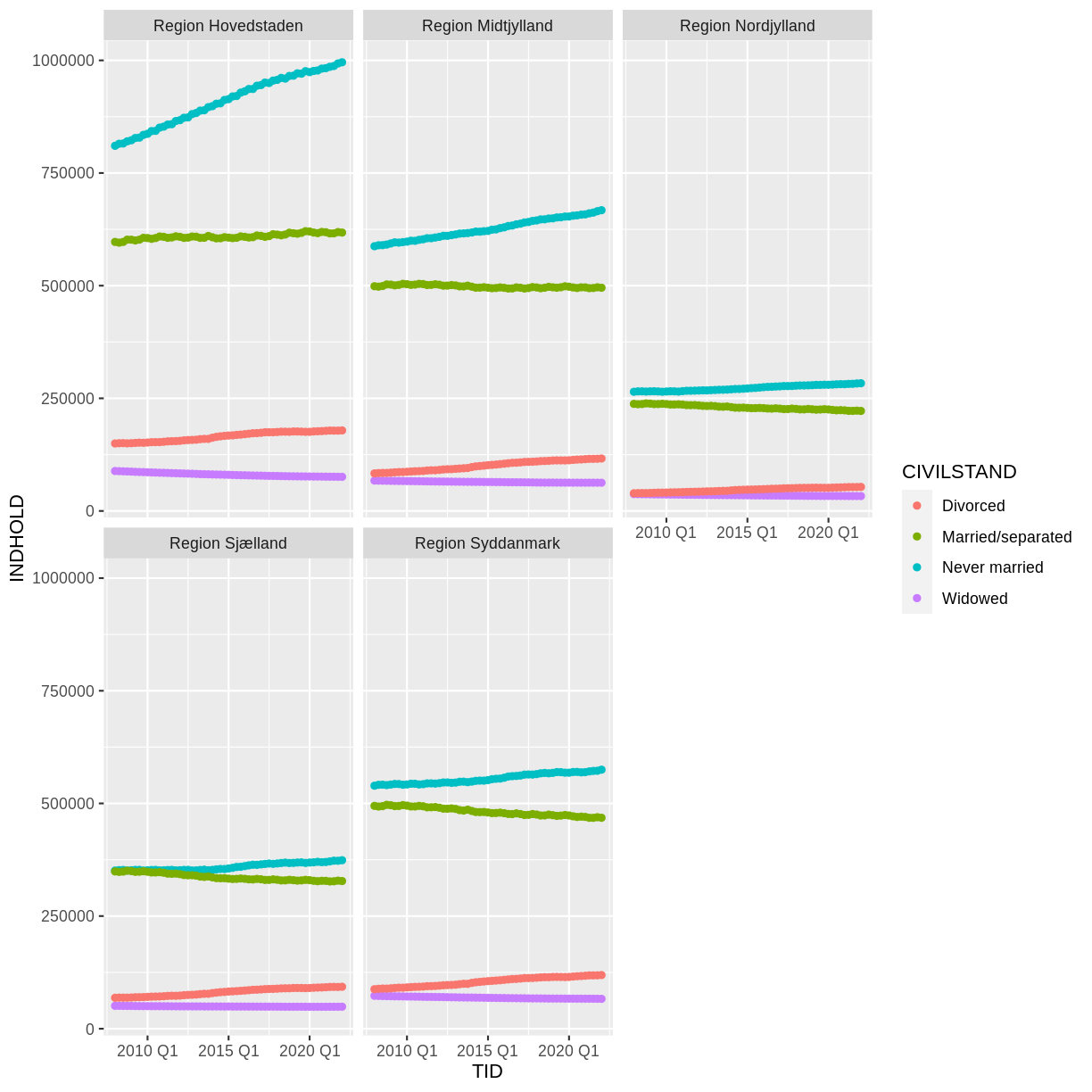
Click the “Zoom” button in your RStudio plots pane to view a larger version of this plot.
Boxplot
We can use boxplots to visualize the distribution of observations for each CIVILSTAND:
plot_data %>%
ggplot(aes(x = CIVILSTAND, y = INDHOLD)) +
geom_boxplot()
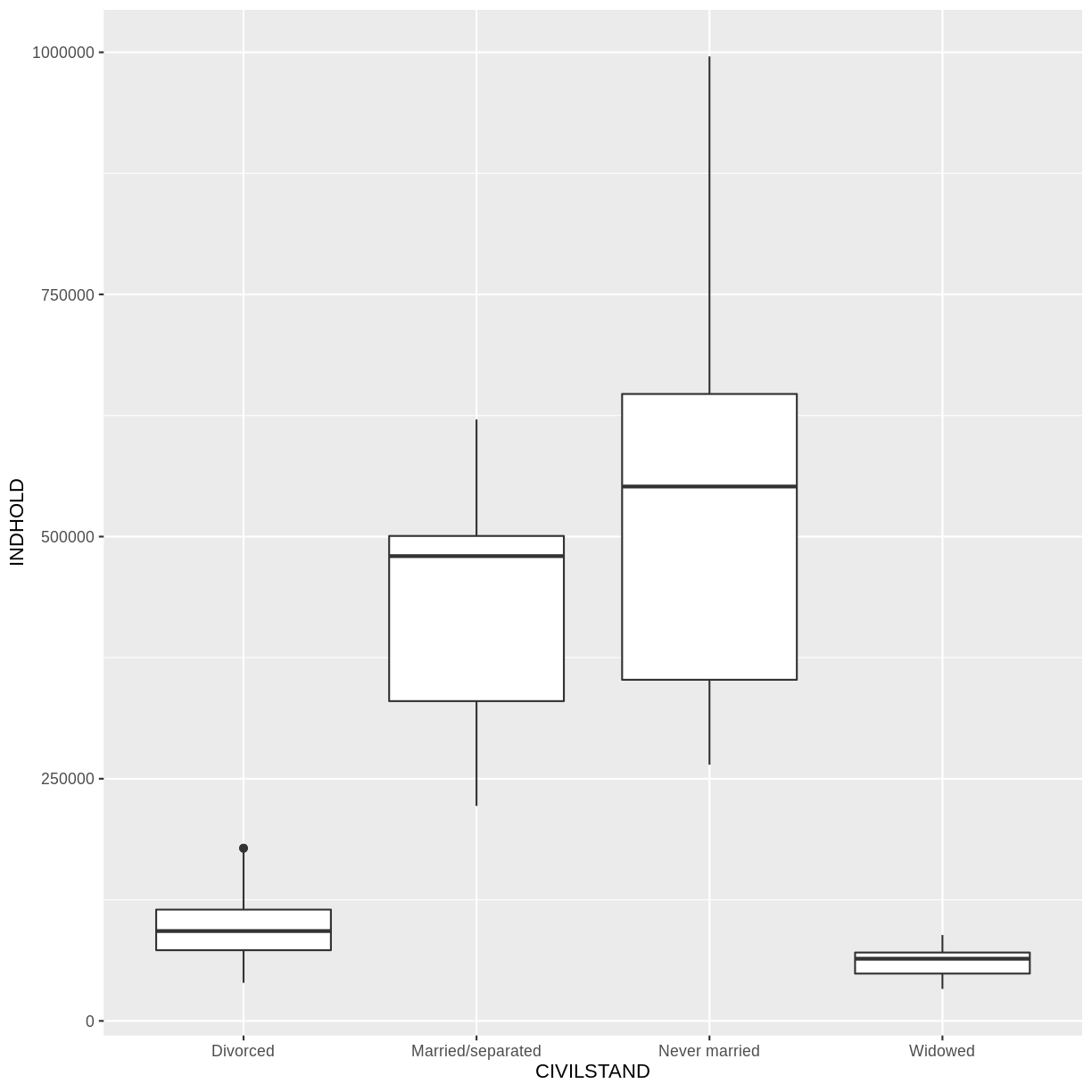 Let us be frank - a boxplot of these aggregated data is not really that
useful. Boxplots are however so useful, that it is relevant to show how they
are made.
Let us be frank - a boxplot of these aggregated data is not really that
useful. Boxplots are however so useful, that it is relevant to show how they
are made.
By adding points to a boxplot, we can have a better idea of the number of measurements and of their distribution:
plot_data %>%
ggplot(aes(x = CIVILSTAND, y = INDHOLD)) +
geom_boxplot() +
geom_jitter(alpha = 0.5,
color = "tomato",
width = 0.2,
height = 0.2)
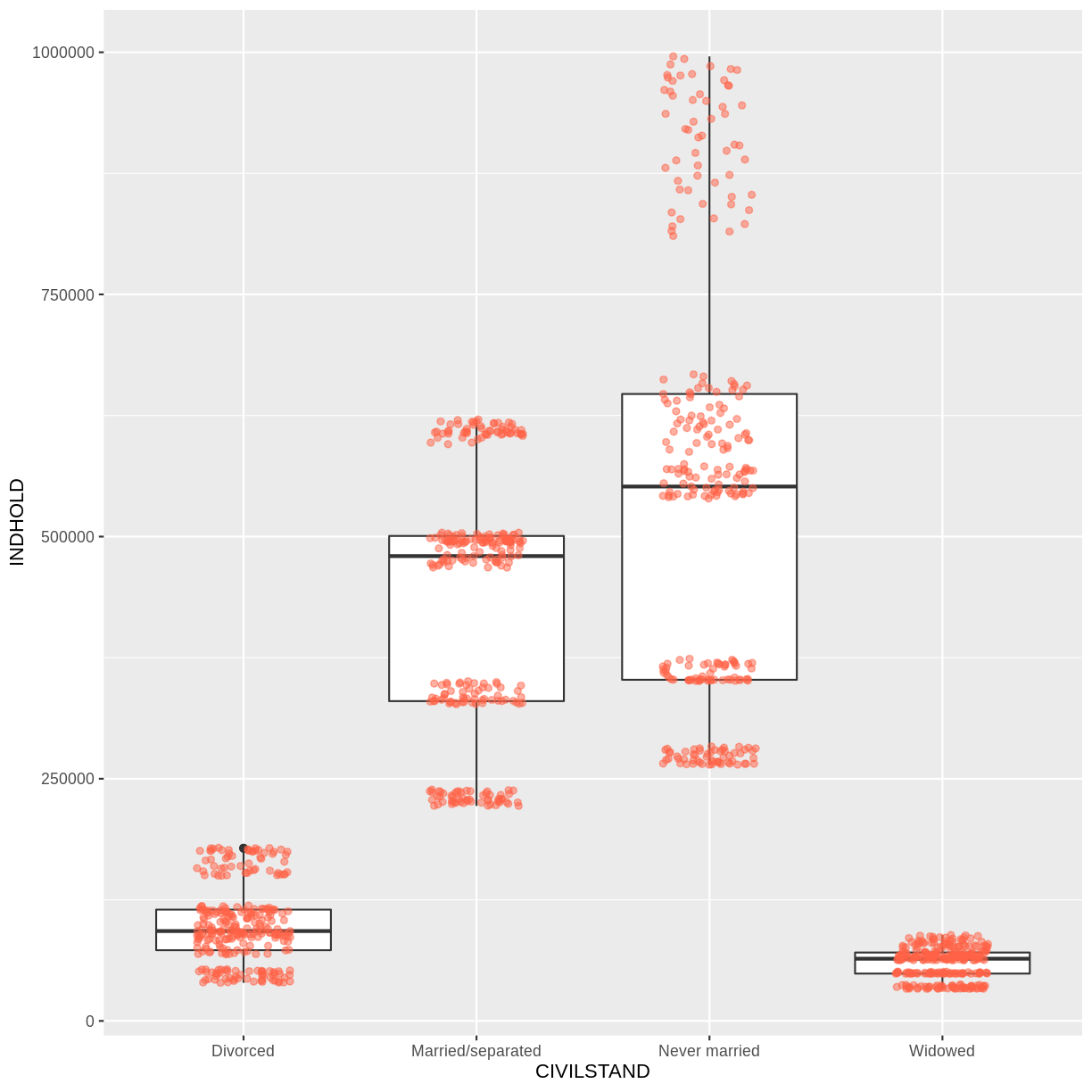 Jitter is a special way of plotting points. When we plot the points at their
exact location, we risk that some of the points overlap. geom_jitter adds a small
bit of noise to the data, in order to spread them out. That way we can better see
individual points.
Jitter is a special way of plotting points. When we plot the points at their
exact location, we risk that some of the points overlap. geom_jitter adds a small
bit of noise to the data, in order to spread them out. That way we can better see
individual points.
Notice how the boxplot layer is behind the jitter layer? What do you need to change in the code to put the boxplot in behind the points such that it’s not hidden?
Barplots
Barplots are also useful for visualizing categorical data. By default,
geom_bar accepts a variable for x, and plots the number of instances each
value of x (in this case, wall type) appears in the dataset.
plot_data %>%
ggplot(aes(x = CIVILSTAND)) +
geom_bar()
We have an equal number of datapoints for each value of “CIVILSTAND”. Not that useful.
Rather than using the default “count” of values, we can use the values directly. In that case, we need to provide both the x- and the y-values; ggplot does not calculate them!
plot_data %>% ggplot(aes(CIVILSTAND, INDHOLD)) +
geom_bar(stat="identity")
 Now we get the values from INDHOLD plotted on the y-axis. But we get ALL the
values from INDHOLD plotted. And we have INDHOLD from several years, from
several administrative parts of Denmark.
Now we get the values from INDHOLD plotted on the y-axis. But we get ALL the
values from INDHOLD plotted. And we have INDHOLD from several years, from
several administrative parts of Denmark.
Let us filter the data.
str_detect(OMRÅDE, “Region”) picks out the rows containing the text “Region”.
TID == yearquarter(“2008 Q1”) picks out the rows containing data from the first quarter of 2008. Note that we have to convert “2008 Q1” to the same datatype as is contained in the columns, using the yearquarter() function.
plot_data %>%
filter(str_detect(OMRÅDE, "Region"),
TID == yearquarter("2008 Q1")) %>%
ggplot(aes(CIVILSTAND, INDHOLD)) +
geom_bar(stat= "identity")
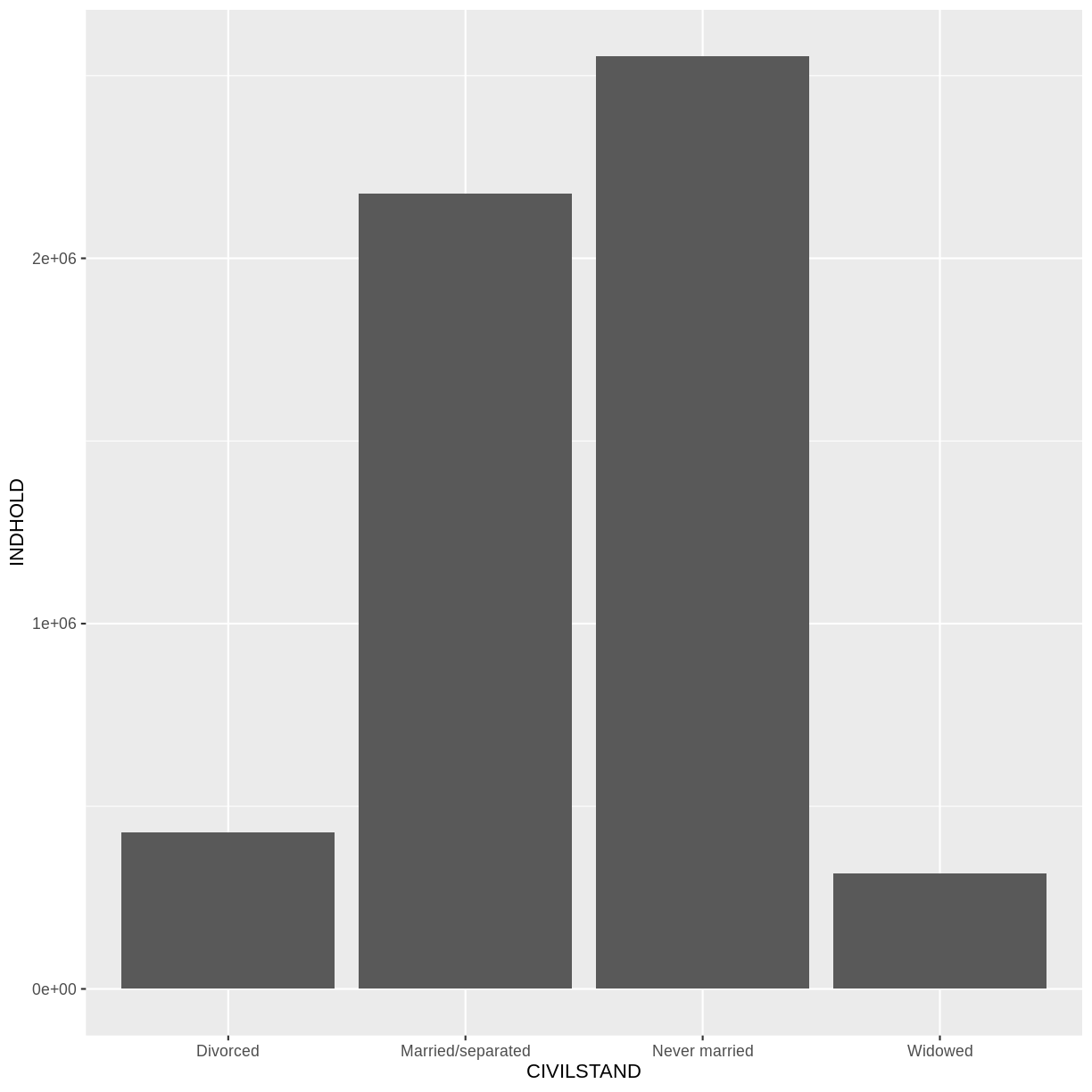 Now we get more sensible numbers. But each bar is still the sum of the number
of divorced persons in ALL the regions.
Now we get more sensible numbers. But each bar is still the sum of the number
of divorced persons in ALL the regions.
We can color bars by region:
plot_data %>%
filter(str_detect(OMRÅDE, "Region"),
TID == yearquarter("2008 Q1")) %>%
ggplot(aes(CIVILSTAND, INDHOLD, color=OMRÅDE)) +
geom_bar(stat= "identity")
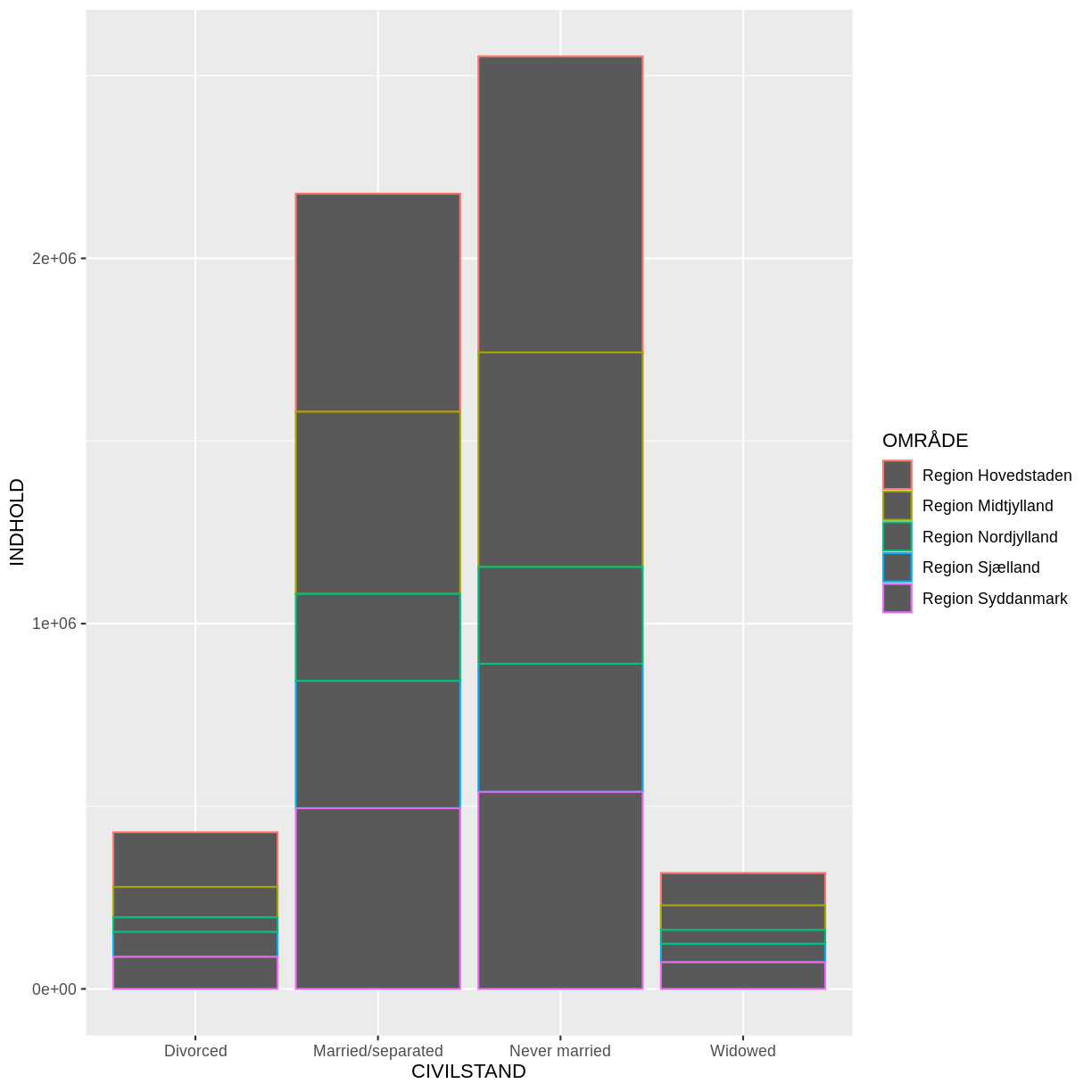 Oops! Color only colors the outline of the bars. We can do better.
Oops! Color only colors the outline of the bars. We can do better.
We can use the fill aesthetic for the geom_bar() geom to colour bars by
the portion of each count that is from each OMRÅDE.
plot_data %>%
filter(str_detect(OMRÅDE, "Region"),
TID == yearquarter("2008 Q1")) %>%
ggplot(aes(CIVILSTAND, INDHOLD, fill=OMRÅDE)) +
geom_bar(stat= "identity")
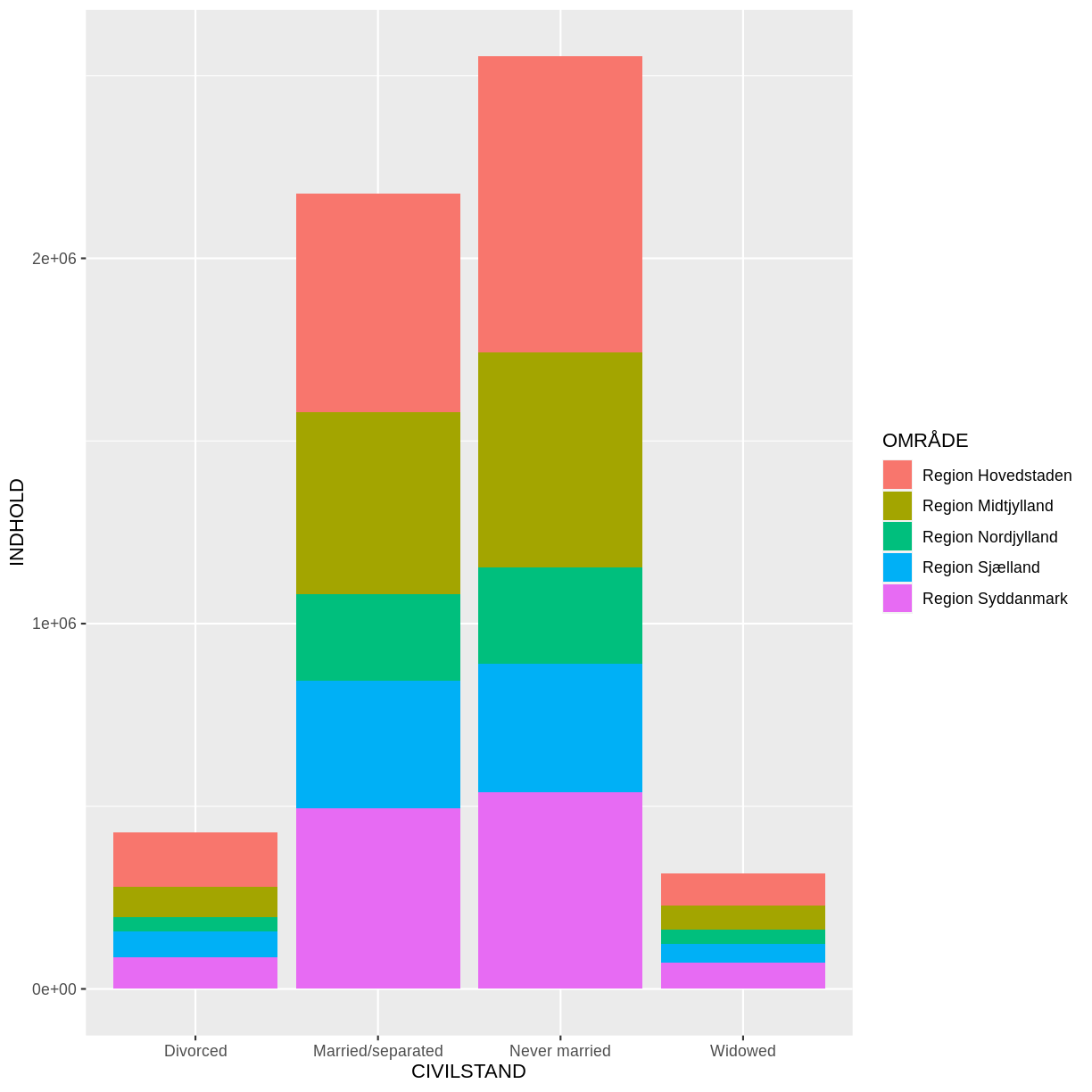
This creates a stacked bar chart. These are generally more difficult to read
than side-by-side bars. We can separate the portions of the stacked bar that
correspond to each OMRÅDE and put them side-by-side by using the position
argument for geom_bar() and setting it to “dodge”.
plot_data %>%
filter(str_detect(OMRÅDE, "Region"),
TID == yearquarter("2008 Q1")) %>%
ggplot(aes(CIVILSTAND, INDHOLD, fill=OMRÅDE)) +
geom_bar(stat= "identity", position = "dodge")
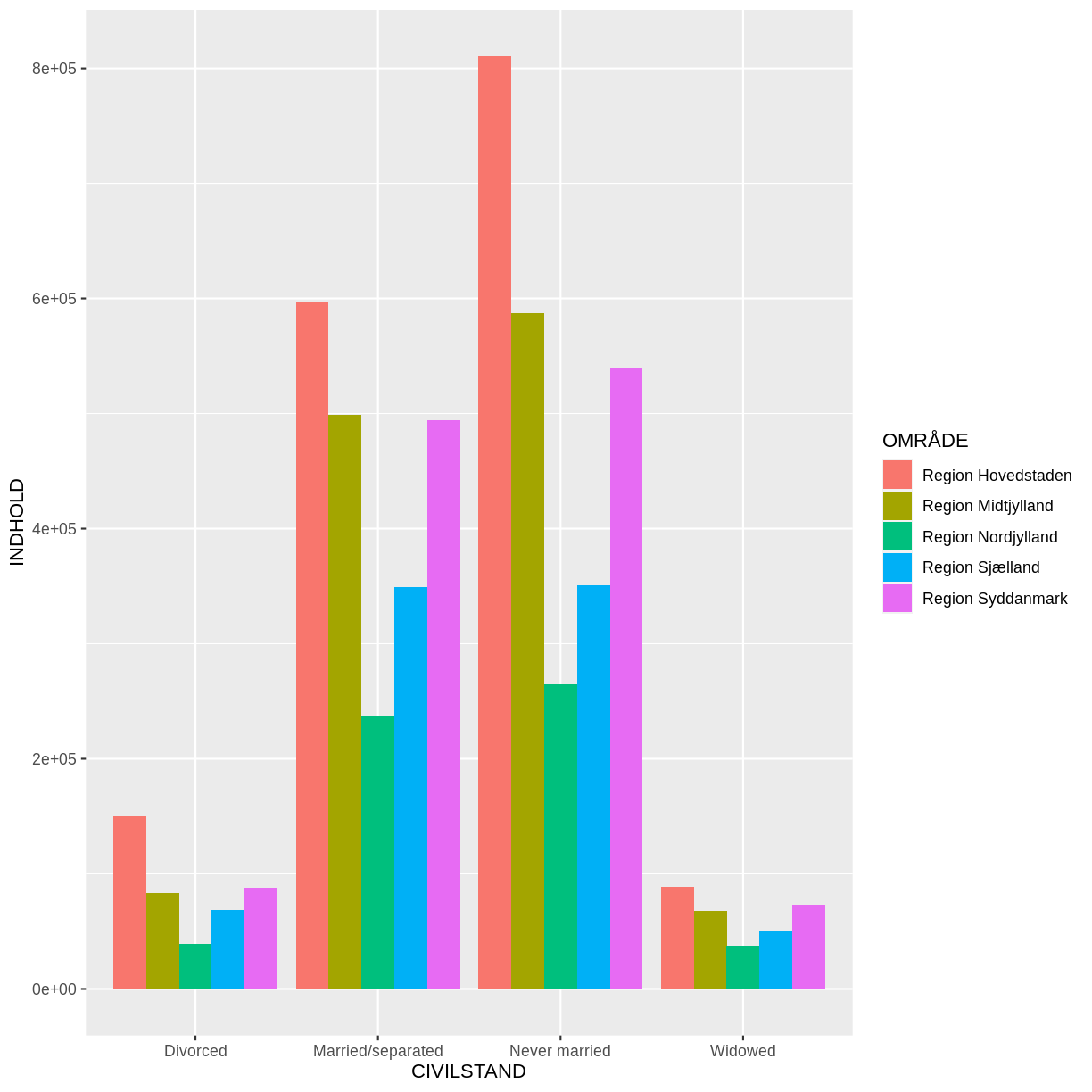
Adding Labels and Titles
By default, the axes labels on a plot are determined by the name of the variable
being plotted. However, ggplot2 offers lots of customization options,
like specifying the axes labels, and adding a title to the plot with
relatively few lines of code. We will add more informative x-and y-axis
labels to our plot, a more explanatory label to the legend, and a plot title.
The labs function takes the following arguments:
title– to produce a plot titlesubtitle– to produce a plot subtitle (smaller text placed beneath the title)caption– a caption for the plot...– any pair of name and value for aesthetics used in the plot (e.g.,x,y,fill,color,size)
plot_data %>%
filter(str_detect(OMRÅDE, "Region"),
TID == yearquarter("2008 Q1")) %>%
ggplot(aes(CIVILSTAND, INDHOLD, fill=OMRÅDE)) +
geom_bar(stat= "identity", position = "dodge") +
labs(title = "Civilstand by region",
subtitle = "First quarter of 2008",
x = "Region",
y = "Number",
caption = "Pattern appears similar between the regions. Data from Statistics Denmark")
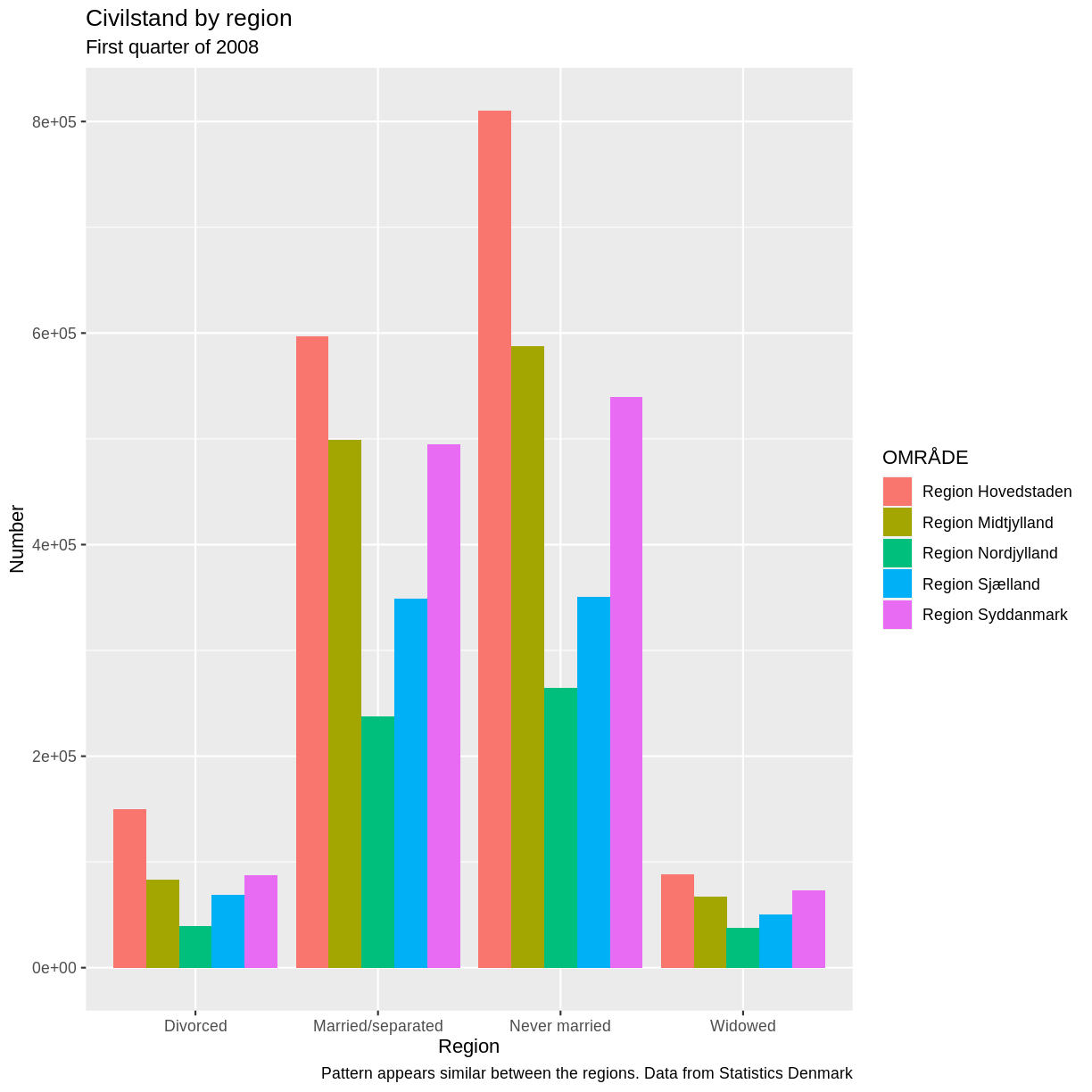
Usually plots with white background look more readable when printed. We can set
the background to white using the function theme_bw(). Additionally, you can remove
the grid:
plot_data %>%
filter(str_detect(OMRÅDE, "Region"),
TID == yearquarter("2008 Q1")) %>%
ggplot(aes(CIVILSTAND, INDHOLD, fill=OMRÅDE)) +
geom_bar(stat= "identity", position = "dodge") +
labs(title = "Civilstand by region",
subtitle = "First quarter of 2008",
x = "Region",
y = "Number",
caption = "Pattern appears similar between the regions. Data from Statistics Denmark") +
theme_bw() +
theme(panel.grid = element_blank())
Key Points
ggplot2is a flexible and useful tool for creating plots in R.The data set and coordinate system can be defined using the
ggplotfunction.Additional layers, including geoms, are added using the
+operator.Boxplots are useful for visualizing the distribution of a continuous variable.
Barplots are useful for visualizing categorical data.
Faceting allows you to generate multiple plots based on a categorical variable.
Whats next?
Overview
Teaching: 30 min
Exercises: 15 minQuestions
What is the next step?
Objectives
Get an idea about what to do to learn more

Key Points
Practice makes perfect
KUB Datalab offers lots of courses and consultations
The web is overflowing with tutorials and courses