A deeper dive into pipes
Last updated on 2026-02-03 | Edit this page
Overview
Questions
- What are the differences between the two pipes?
Objectives
- Explain some of the differences between the two families of pipes
Almost everywhere in this course material, you will see the pipe.
But the story about the pipe is larger than %>%.
The %>% version come from the magrittr
package, and was introduced to the R-world in 2014. It became one of the
defining features of the R-dialect “tidyverse”. It proved useful enough
that a similar pipe |> was introduced in “base-R” with
version 4.1 in May 2021.
R-programmers tend to write in one dialect of R, small variations that are understandable - but still different. The three common dialects are “base R”, the tidyverse and data.table. One of the defining features of the tidyverse dialect has been the magrittr pipe. This differences is eroding with the introduction of the native pipe in base R.
KUB Datalab speaks the tidyverse dialect, and will continue to use the magrittr pipe until the default keyboard shortcut to the pipe in RStudio changes
The base-R pipe function in a similar way to the magrittr pipe. But there are differences:
| Topic | Magrittr 2.0.3 | Base 4.3.0 | note |
|---|---|---|---|
| Operator | %\>% %\<\>% %$% %!>% %T>% |
|> |
1 |
| Function calls | 1:2 %>% sum() |
1:2 |> sum() |
|
1:2 %>% sum |
Needs the parentheses | ||
1:3 %>%`+` 4
|
Not every function is supported | ||
| Insert on first empty place | mtcars %>% lm(formula=mpg ~ disp) |
mtcars |> lm(formula = mpg ~ disp) |
2 |
| placeholder | . | _ | |
mtcars %>% lm(mpg~disp, data = .) |
mtcars |> lm(mpg ~ disp, data = _) |
||
mtcars %>% lm(mpg~disp, .) |
Needs named argument | 3 | |
1:3 %>% setNames(., .) |
Can only appear once | ||
1:3 %>% {sum(sqrt(.))} |
Nested calls not allowed | 4 | |
| Extraction call |
|
|
|
| Environment |
%>% has additional function environment. Use:
"x" %!> assign(1)
|
"x" |> assign(1) |
4 |
| Create Function | top6 <- . %>% sort() %>% tail() |
Not possible | |
| Speed | Slower because overhead of function call | Faster because syntax transformation | 5 |
- Unless we explicitly import the
magrittrpackage, we typically only have access to the simple pipe,%>%which is included in many packages. This however is only one of several pipes available from themagrittrpackage. See below. - Note that both pipes insert the input from the left-hand-side at the first unnamed argument. Naming other arguments allow us to handle functions that do not take data as the first argument.
- The placeholder allow us to reference the input in functions. One important difference is that the base-R pipe need to be specified with a named argument. Which is probably best practice anyway.
- This is related to the way the pipes actually work, see also 5.
- The magrittr pipe works by rewriting the right-hand-side in a function, with its own environment - where the left-hand-side is available with the symbol `.`
The other pipes
As indicated in the comparative table, the magrittr pipe
have a number of siblings.
The tee-pipe %T>%
This pipe allow us to inject a function into a longer set of pipes, without affecting the rest of the flow.

The tee-pipe sends data to two functions. The immediate next, which we want to run for plotting, logging or printing data, and to the next function over, which get the original input data, rather than what is returned by the intermediate function.
We are here using an intermediate function, not for what it does to the data, but for its side effects.
Try it yourself
Use the tee-pipe to send the dataset mtcars to a view
function, and then filter to only show rows where am is
equal to 1.
Assignment pipen
After constructing a longer pipe in order to get at specific result, we end up assigning the result to a new object. Sometimes the same object:
R
my_data <- my_data |>
filter(lending > 10) |>
mutate(area = lenght * width)
The assignment pipe can shorten this:
R
my_data %<>%
filter(lending > 10) |>
mutate(area = lenght * width)
It needs to be the first pipe in the flow, and will pass the data from its left hand side to the flow on the right hand side. And assign the result of the entire pipe back into the original left hand side.
This will shorten your code. But it is generally recommended not to use it.
Reading R-code can be difficult enough as it is, and
%>% is visually rather close to %<>%
which makes it easy to overlook.
The eager pipe %!>%
We definitely need an episode in this material that cover the concept
of lazy evaluation. But currently we are too lazy to write
it.
But. The usual pipe %>% i “lazy”. An expression in a
set of pipes is only evaluated when R absolutely have to. This can do
weird things, especially if we run functions for their side effects,
e.g. when we use the tee-pipe to do logging of intermediate results.
Enter the eager pipe, %!>%. This will force the
evaluation rather than postpone it to later.
Let us define three functions. The code is not that important, but they take an input, prints a text indicating their name (ie function f prints “function f”), and return the input unchanged. This printing is the side effect.
Take a look below if you are curious:
R
f <- function(x) {
print("function f")
x
}
g <- function(x) {
print("function g")
x
}
h <- function(x) {
print("function h")
invisible(x)
}
invisible() returns x, but withou printing it.
We can now run these three functions in a pipe:
R
library(magrittr)
NULL %>% f() %>% g() %>% h()
OUTPUT
[1] "function h"
[1] "function g"
[1] "function f"This is the lazy evaluation. The first print come from the last function, and function g is only evaluated when it has to be. Because function h needs it. The last function to be evaluated is function f - the first we actually have in the pipes. Because it is only evaluated when needed. That is, when function g needs it.
Eager evaluation, where the functions are not evaluated when needed, but immediately, can be achieved using the eager pipe:
R
NULL %!>% f() %!>% g() %!>% h()
OUTPUT
[1] "function f"
[1] "function g"
[1] "function h"Now the first message printed is from function f. Becaus function f is the first, and we are using the eager pipe rather than the lazy.
R use lazy evaluation for a reason
It might be tempting to use the eager pipe every time. After all, it is bad to be lazy and the evaluation have to be done at one point, right. But! R, and especially the tidyverse functions use lazy evaluation because it improves performance. If something in the pipe is not actually used, it is not evaluated. This saves time.
The exposition pipe
The power of the pipes comes from the fact that we can use them to forward data to a function that use a dataframe.
But not all functions have an argument that allow them to take a dataframe as input. And because the pipe inserts whatever is on the lefthand side of it, into the first empty argument in the function on the right hand side, this can lead to problems.
Try it yourself
You will fail!
Use the cor() function in conjunction with the ordinary
pipe (%>%), to find the correlation between the Sepal.Length and the
Sepal.Width in the iris dataset.
Something like this:
R
iris %>%
cor(x = .$Sepal.Length, y = .$Sepal.Width)
Might be tempting. Try to add other named arguments to the function (look at the help file) if you do not succeed at first.
This cannot be done.
In this attempt:
R
iris %>%
cor(x = .$Sepal.Length, y = .$Sepal.Width,
use = "everything")
We add more and more named arguments to the function. In this code,
we will get an error, telling us that we are supplying a dataframe to
the argument “method”. Looking at the help, we can see that
cor besides the arguments we have already named in the
function call, take a final argument “method”. That is the first empty
argument in the function, and the pipe will place the dataframe
iris on that place, leading to an error. If we supply the
method argument inserting method = "pearson", the pipe will
still add the dataframe to the function, resulting in an error telling
us that we are supplying an argument that is unused.
The exposition pipe, %$% exposes the variables in the
left hand side, and does not insert anything anywhere unless we specify
it:
R
iris %$%
cor(x = Sepal.Length, y = Sepal.Width)
OUTPUT
[1] -0.1175698That weird note about function calls
In the table comparing the native pipe (|>) and the
magrittr pipe (%>%) it was noted that the latter was
slower than the first, because of “overhead of function call”. Or
conversely that the first was faster because of “syntax transformation”.
What does that actually mean?
The magrittr pipe, as are other operators, is a function.
Conceptually this a %>% b(), is the same as
%>%(a, b())
Or:
R
data %>%
filter(x > 0) %>%
summarise(mean = mean(y))
become this:
R
`%>%`(`%>%`(data, filter(x > 0)), summarize(mean = mean(y)))
We can count 5 functions, 2 of them the pipe function.
The native pipe on the other hand is not a function. The R-parser,
which interprets our code befor actually evaluating it, rewrites code
containg the native pipe. That means, that a |> b()
becomes b(a)
Or that:
R
data |>
filter(x > 0) |>
summarize(mean = mean(y))
become:
R
summarize(filter(data, x > 0), mean = mean(y))
We can count 3 functions in this expression. Intuitively this should be faster. And it is.
What if I want the other pipe?
The keyboard shortcut to get a pipe operator in RStudio is ctrl+shift+m on windows and cmd+shift+m on macos.
This default can be changed:
Tools → Global Options → Code
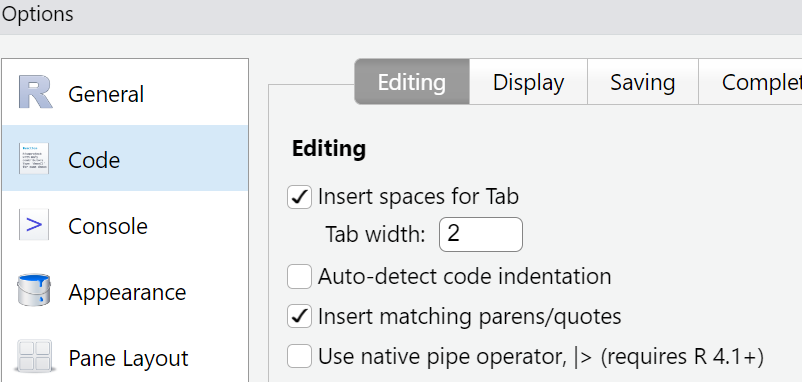 Place a tickmark in the box next to
“Use native pipe operator, |>”.
Place a tickmark in the box next to
“Use native pipe operator, |>”.
- The base-R, native, pipe |> is faster than the magrittr pipe, %>%
- The magrittr package provides a selection of other useful pipes
